QTL analysis for Crichton final
Hao He
Last updated: 2023-02-04
Checks: 7 0
Knit directory: QTL_analysis_for_Crichton/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221224) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ff93680. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: output/WT144_effect_for_marker__int_chr5-136332619-.-C-A.pdf
Untracked: output/WT144_effect_for_marker_plots_TotalDist_vs_TestAge_by_marker.pdf
Untracked: output/WT144_plot-Intercept.pdf
Untracked: output/WT144_plot-Slope.pdf
Untracked: output/WT144_plot-TestAge.x.pdf
Untracked: output/WT144_plot-TestAge.y.pdf
Untracked: output/WT144_plot-TotalDist_1.pdf
Untracked: output/WT144_plot-TotalDist_2.pdf
Untracked: output/WT144_plot-TotalDist_3.pdf
Untracked: output/WT144_plot-base.x.pdf
Untracked: output/WT144_plot-base.y.pdf
Untracked: output/figure_panel.pdf
Untracked: output/qtl.out.obj.mixedmodel.RData
Untracked: output/qtl.out.obj.mixedmodel.final.RData
Unstaged changes:
Modified: analysis/workflow_proc.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/QTL_analysis_for_Crichton_final.Rmd) and HTML (docs/QTL_analysis_for_Crichton_final.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | ff93680 | xhyuo | 2023-02-04 | change figure panel |
| html | 2470aab | xhyuo | 2022-12-24 | Build site. |
| Rmd | 92ecd1d | xhyuo | 2022-12-24 | First build |
Library
library(ggplot2)
library(gridExtra)
library(qtl)
library(qtlcharts)
library(tidyverse)
library(lme4)
library(lmerTest)
library(qtl2)
library(cowplot)Load qtl data and plot summary
WT144 <- read.cross(format = "csv", file = "data/WT144_GBS_Rqtl_input.csv", genotypes = c("A", "H", "B")) --Read the following data:
309 individuals
225 markers
11 phenotypes
--Cross type: f2 chror <- as.character(1:19)
WT144$geno <- WT144$geno[chror]
summary(WT144) F2 intercross
No. individuals: 309
No. phenotypes: 11
Percent phenotyped: 100 100 100 99.7 100 100 99.7 90.9 100 99.7 90.9
No. chromosomes: 19
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
Total markers: 225
No. markers: 18 23 17 15 46 1 19 4 8 3 5 18 11 10 2 4 7 2 12
Percent genotyped: 85
Genotypes (%): AA:40.6 AB:46.9 BB:12.5 not BB:0.0 not AA:0.0 plotMissing(WT144, main="")
#drop.nullmarkers
WT144 <- drop.nullmarkers(WT144)
plotMissing(WT144, main="")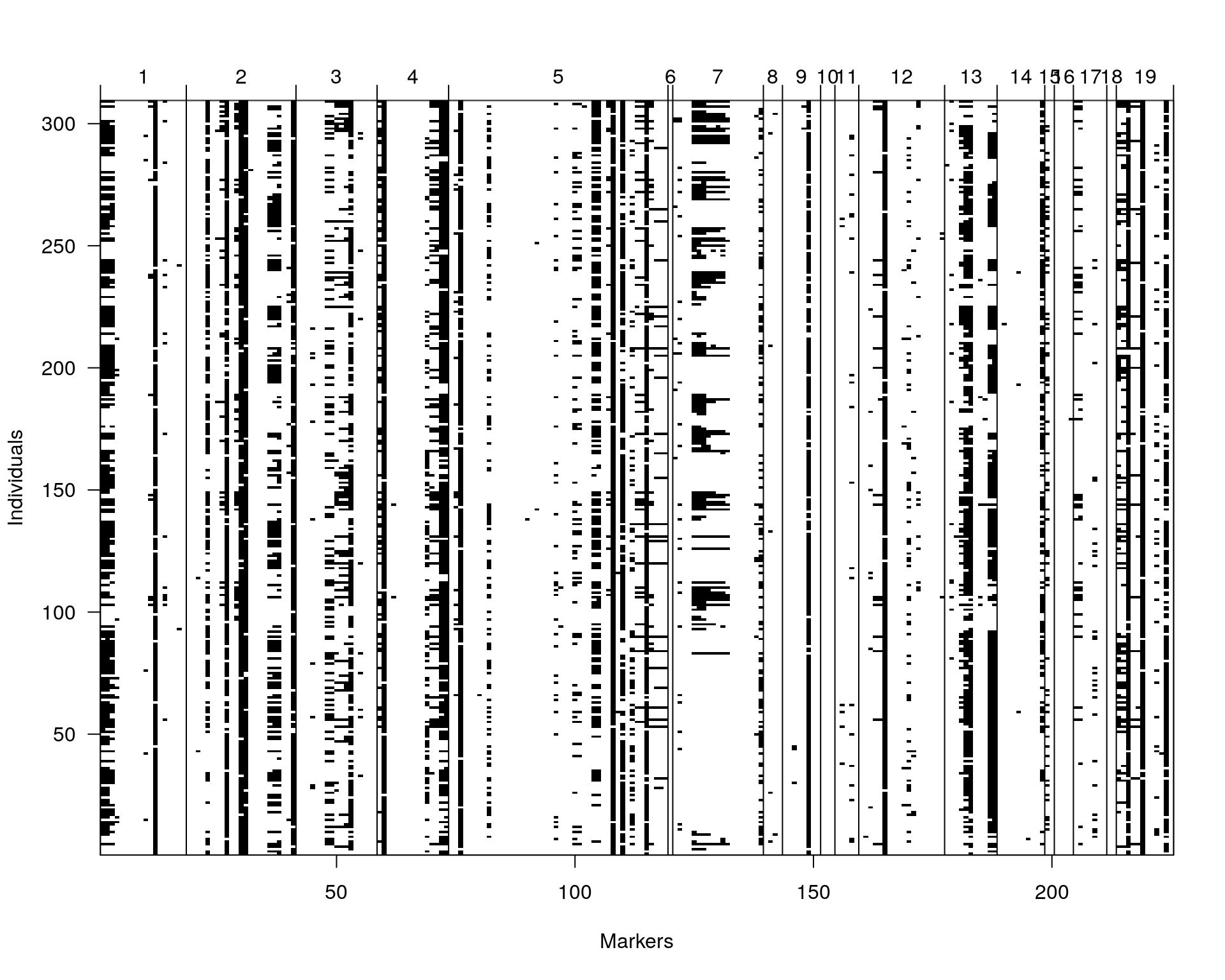
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
Genotype_distribution_pre
# Plot the distribution of homo/hetero for each marker
af <- NULL
for (chr in names(WT144$geno)){
af <- rbind(af, data.frame(chr=chr,
A=colSums(WT144$geno[[chr]]$data == 1, na.rm = T),
H=colSums(WT144$geno[[chr]]$data == 2, na.rm = T),
B=colSums(WT144$geno[[chr]]$data == 3, na.rm = T),
N=colSums(is.na(WT144$geno[[chr]]$data))))
}
af$chr <- factor(af$chr, levels = names(WT144$geno))
af$ID <- 1:nrow(af)
#plot
genotype_distribution_pre <- ggplot(af) +
geom_point(aes(ID, A), color = "red") +
geom_point(aes(ID, B), color="purple") +
geom_point(aes(ID, H), color="green") +
geom_point(aes(ID, N), color="black") +
facet_grid(~chr, scales = "free") +
ylab("Frequency") +
theme(text = element_text(size = 14),
axis.text.x = element_blank(),
axis.ticks = element_blank())
#
print(genotype_distribution_pre)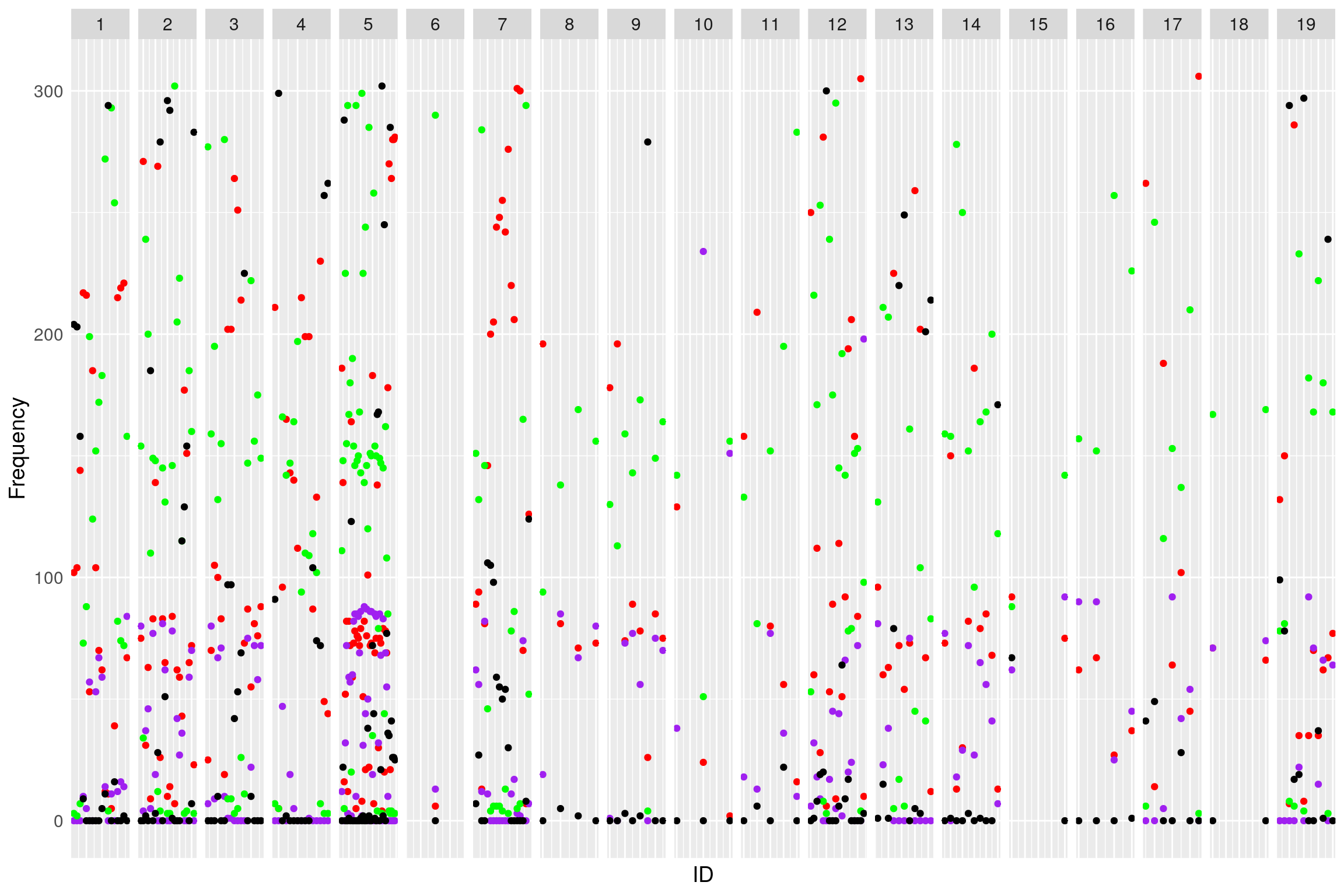
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
Genotype_distribution_post
# Choose who to drop. I chose A > H/4, B > H/4, H > (A+B)/2
dropm <- rownames(af)[af$A < af$H/4 | af$B < af$H/4 | af$H < (af$A+af$B)/2 | af$N > (af$A+af$B+af$H)]
WT144 <- drop.markers(WT144, dropm)
# Plot again
af <- NULL
for (chr in names(WT144$geno)){
af <- rbind(af, data.frame(chr=chr,
A=colSums(WT144$geno[[chr]]$data == 1, na.rm = T),
H=colSums(WT144$geno[[chr]]$data == 2, na.rm = T),
B=colSums(WT144$geno[[chr]]$data == 3, na.rm = T),
N=colSums(is.na(WT144$geno[[chr]]$data))))
}
af$chr <- factor(af$chr, levels = names(WT144$geno))
af$ID <- 1:nrow(af)
#plot
genotype_distribution_post <- ggplot(af) +
geom_point(aes(ID, A), color = "red") +
geom_point(aes(ID, B), color="purple") +
geom_point(aes(ID, H), color="green") +
geom_point(aes(ID, N), color="black") +
facet_grid(~chr, scales = "free") +
ylab("Frequency") +
theme(text = element_text(size = 14),
axis.text.x = element_blank(),
axis.ticks = element_blank())
print(genotype_distribution_post)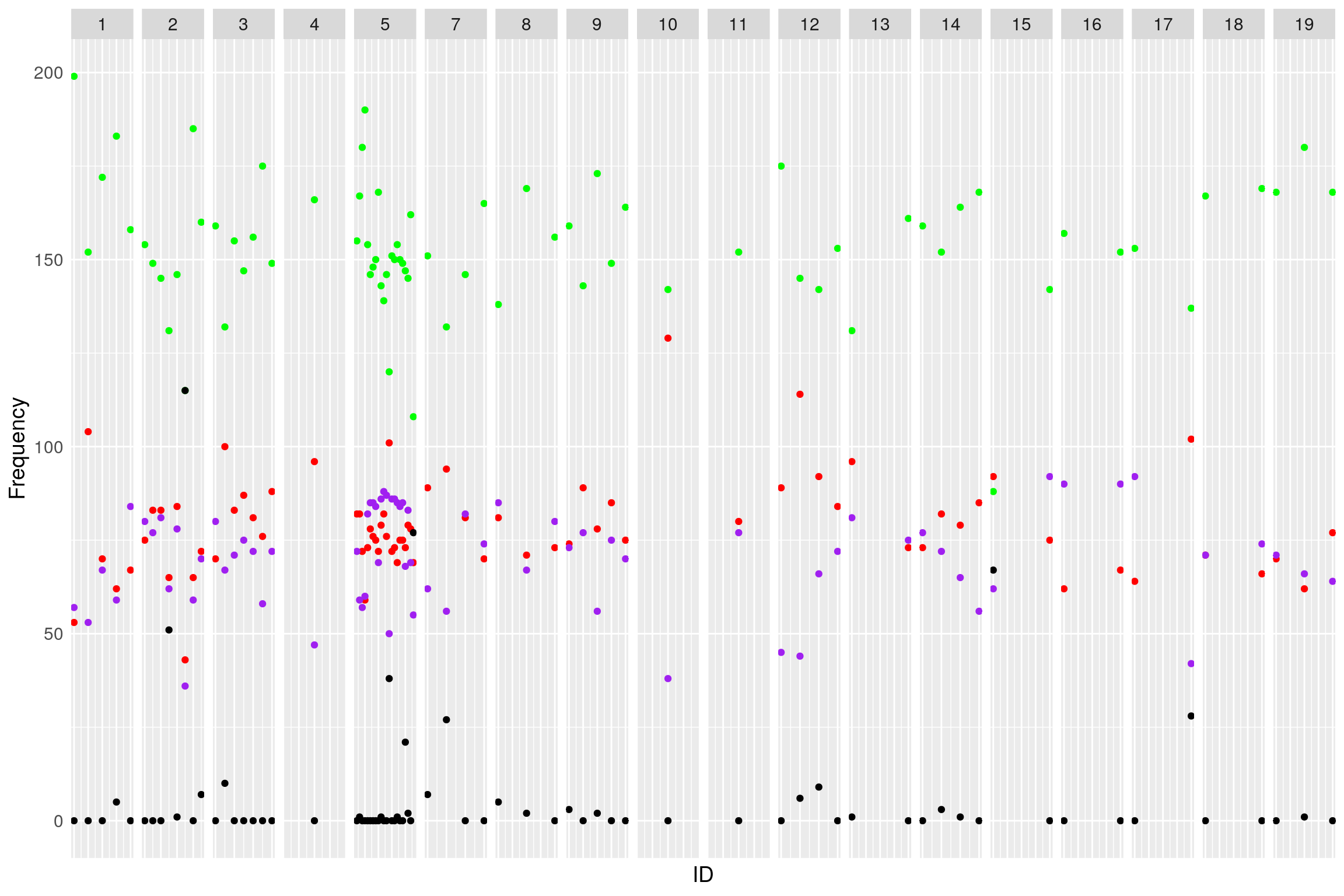
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
plotMap(WT144)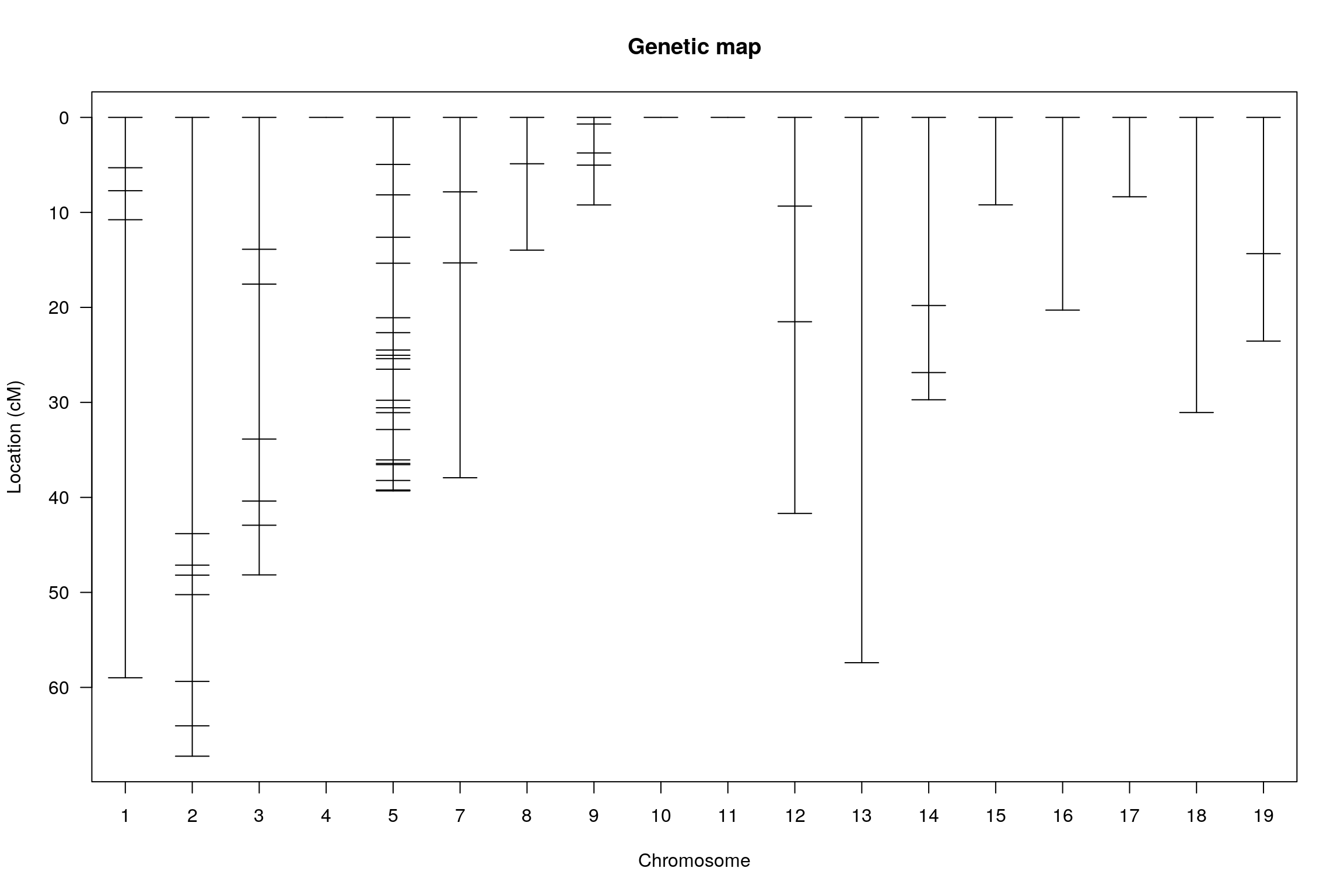
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
Process phenotype on the genotyped animals
#read data Report-12-27-2019.csv and filter to the 309 animals which also have genotypes.
report <- readr::read_csv("data/Report-12-27-2019.csv") %>%
dplyr::select(animal_name, gender, TotalDist, TestAge, test.no) %>%
dplyr::filter(animal_name %in% WT144$pheno$AnimalName) %>% #filter to the animals with genotypes
dplyr::mutate(across(c(gender, test.no), as.factor))
#plot TotalDist vs test age among 309 animals
p6 <- ggplot(data = report,
mapping = aes(x = TestAge, y = TotalDist, color = test.no)) +
geom_point(aes(shape=gender)) +
geom_smooth(method=lm)
print(p6)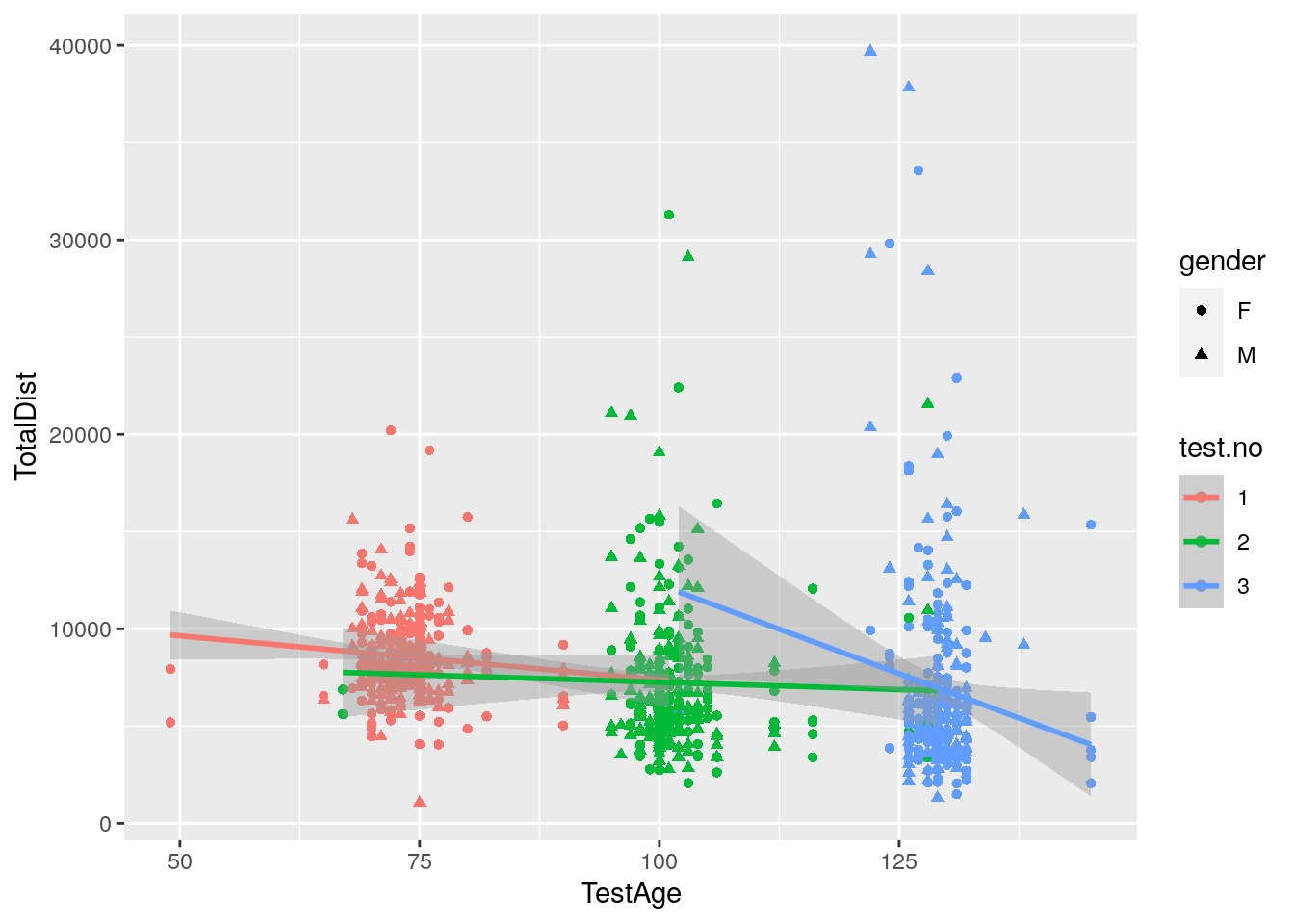
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#plot TotalDist distribution across test.no among 309 animals
p7 <- ggplot(data = report,
mapping = aes(x = test.no, y = TotalDist, color = test.no)) +
geom_boxplot()
print(p7)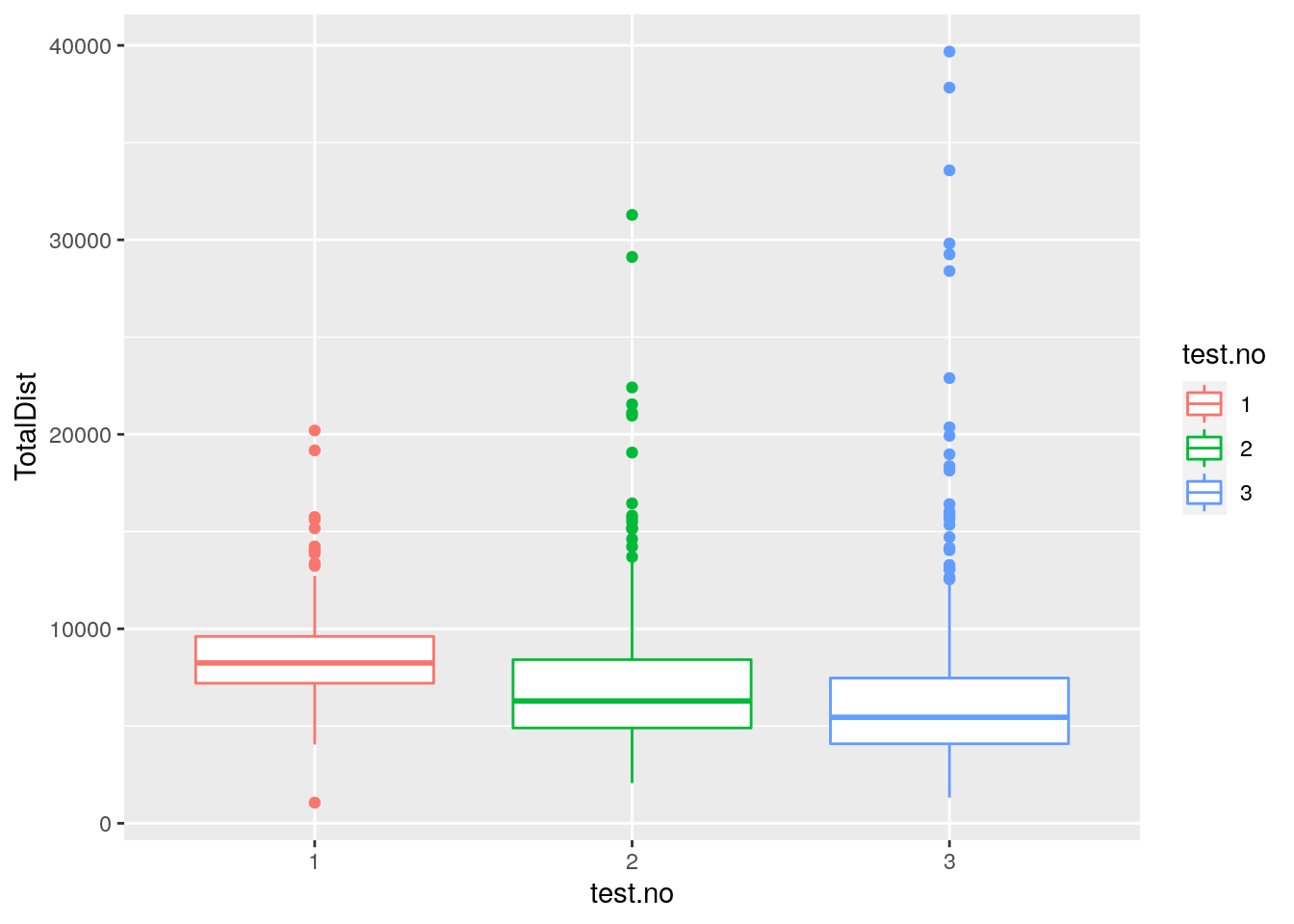
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#plot histgram for TotalDist among 309 animals
p8 <- ggplot(data = report,
mapping = aes(x = TotalDist, fill = test.no)) +
geom_histogram() +
facet_grid(test.no ~ .)
print(p8)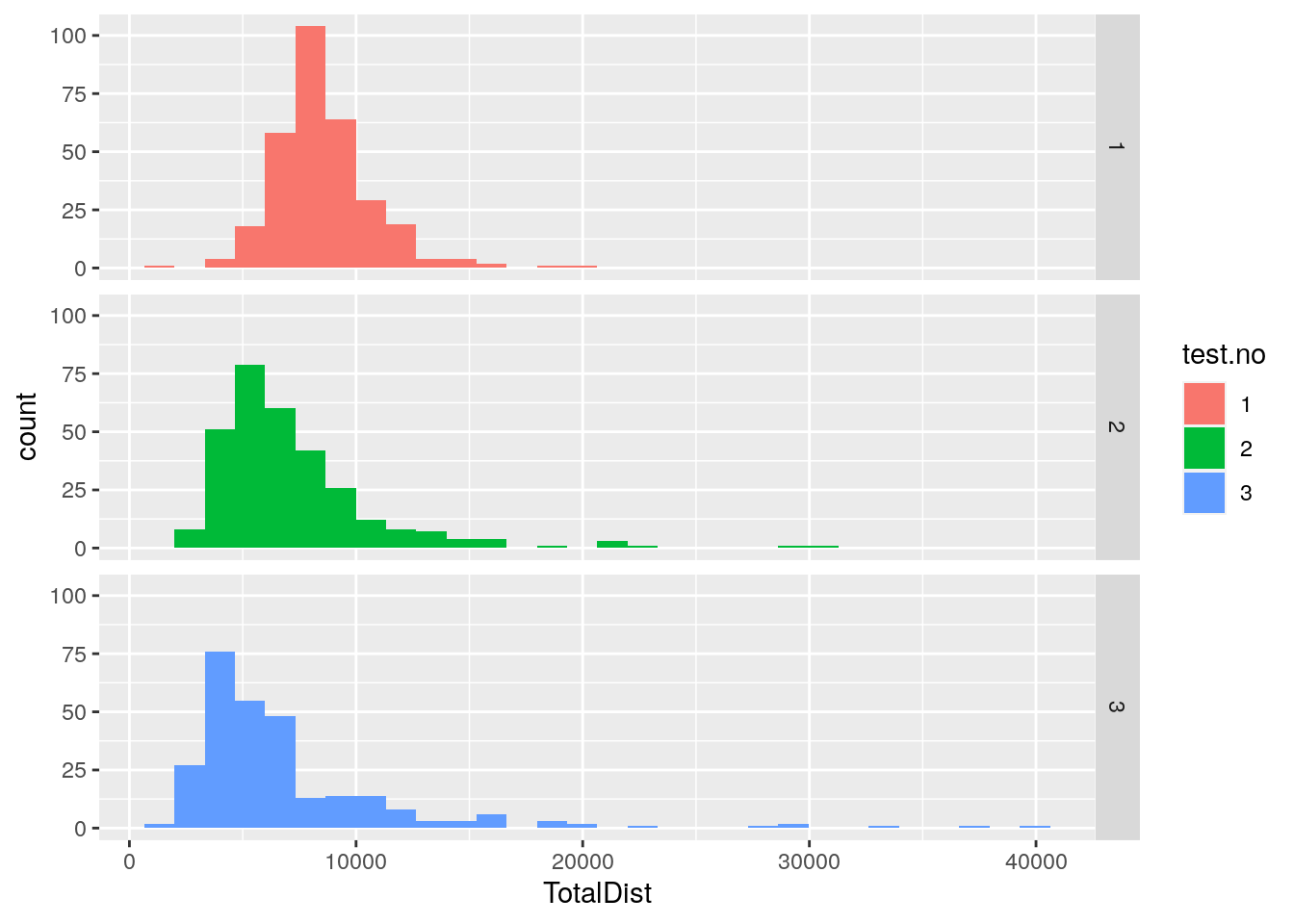
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#plot histgram for TotalDist for genders with mean lines among 309 animals
p9 <- ggplot(data = report,
mapping = aes(x = TotalDist, fill = gender)) +
geom_histogram()
print(p9)
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#a random intercept for each animal,
#and a random slope of TestAge for each animal
#This allows both the intercept and the slope of TestAge to vary across animals
res <- lmer(TotalDist ~ TestAge + (1 + TestAge|animal_name),
data = report)
summary(res)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: TotalDist ~ TestAge + (1 + TestAge | animal_name)
Data: report
REML criterion at convergence: 16596.9
Scaled residuals:
Min 1Q Median 3Q Max
-3.5368 -0.4085 -0.0295 0.3694 3.7790
Random effects:
Groups Name Variance Std.Dev. Corr
animal_name (Intercept) 12321442 3510.2
TestAge 4265 65.3 -0.96
Residual 2173666 1474.3
Number of obs: 898, groups: animal_name, 309
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 10338.640 302.752 321.518 34.149 < 2e-16 ***
TestAge -26.522 4.333 317.190 -6.121 2.75e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr)
TestAge -0.918
optimizer (nloptwrap) convergence code: 0 (OK)
Model failed to converge with max|grad| = 1.58588 (tol = 0.002, component 1)
Model is nearly unidentifiable: very large eigenvalue
- Rescale variables?confint(res, level = 0.95, method = "Wald") 2.5 % 97.5 %
.sig01 NA NA
.sig02 NA NA
.sig03 NA NA
.sigma NA NA
(Intercept) 9745.25700 10932.02232
TestAge -35.01462 -18.02879#there is strong evidence that TestAge significantly decreased TotalDist.
res.std <- resid(res)/sd(resid(res))
plot(res.std, ylab="Standardized Residuals")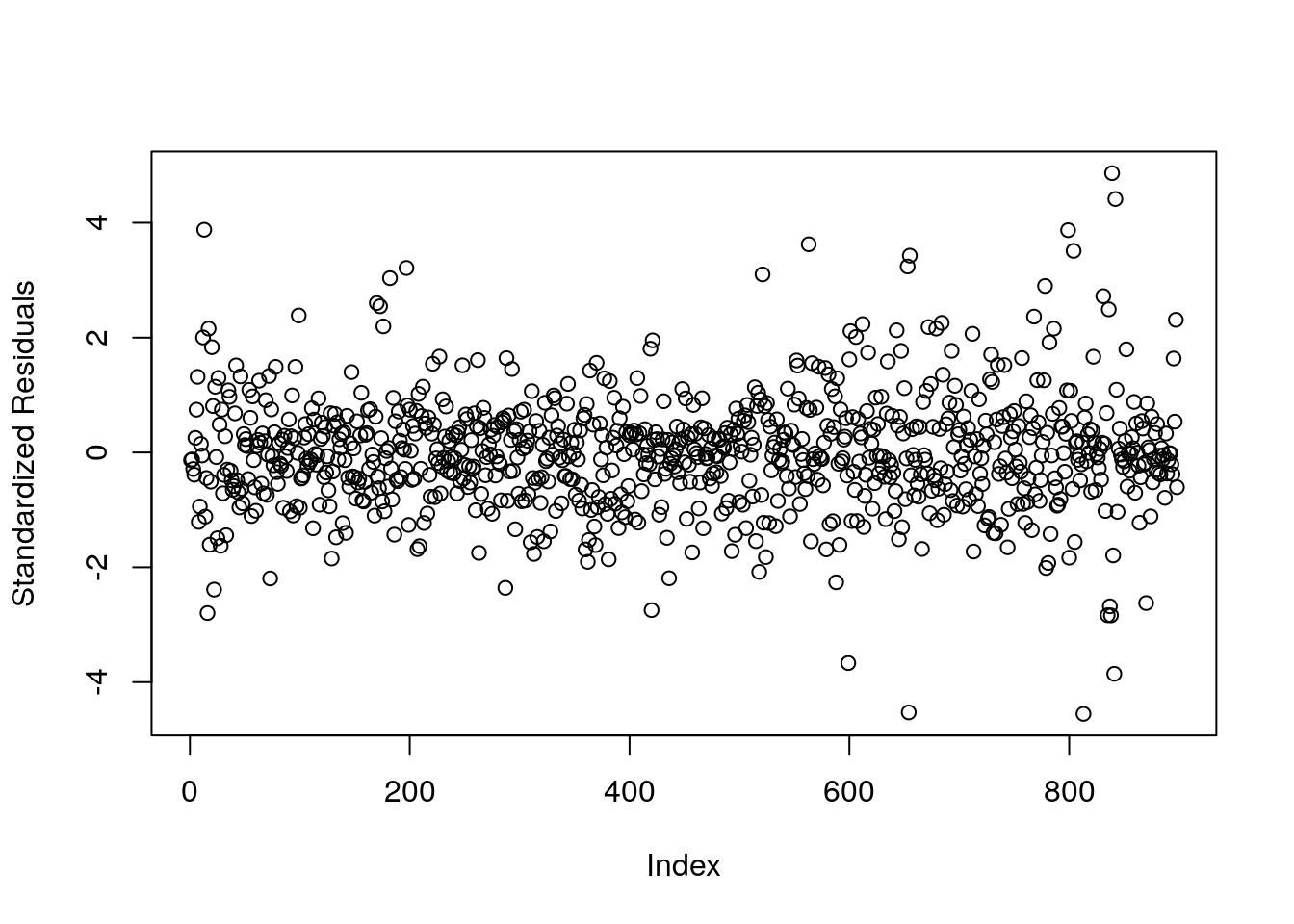
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
ggplot(as.data.frame(res.std), aes(sample = res.std)) +
geom_qq() +
geom_qq_line()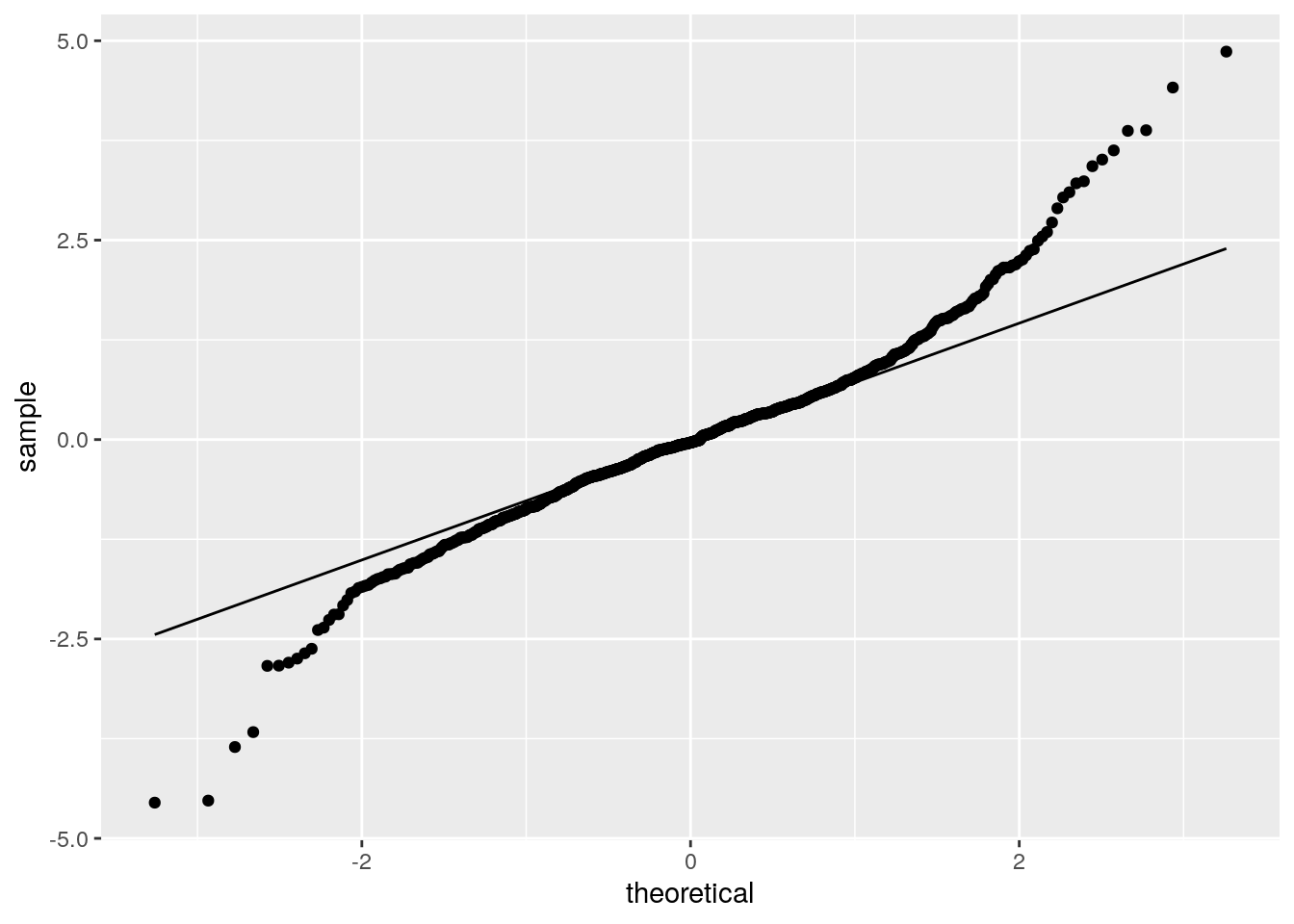
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#to get the fitted Intercept and Slope
TotalDist_animal <- coef(res)$animal_name
colnames(TotalDist_animal) <- c("Intercept", "Slope")
TotalDist_animal$AnimalName <- rownames(TotalDist_animal)
#add TotalDist_animal to the phenotype df in WT144
WT144$pheno <- left_join(WT144$pheno, TotalDist_animal, by = "AnimalName")
plotPheno(WT144, pheno.col=12, xlab = "")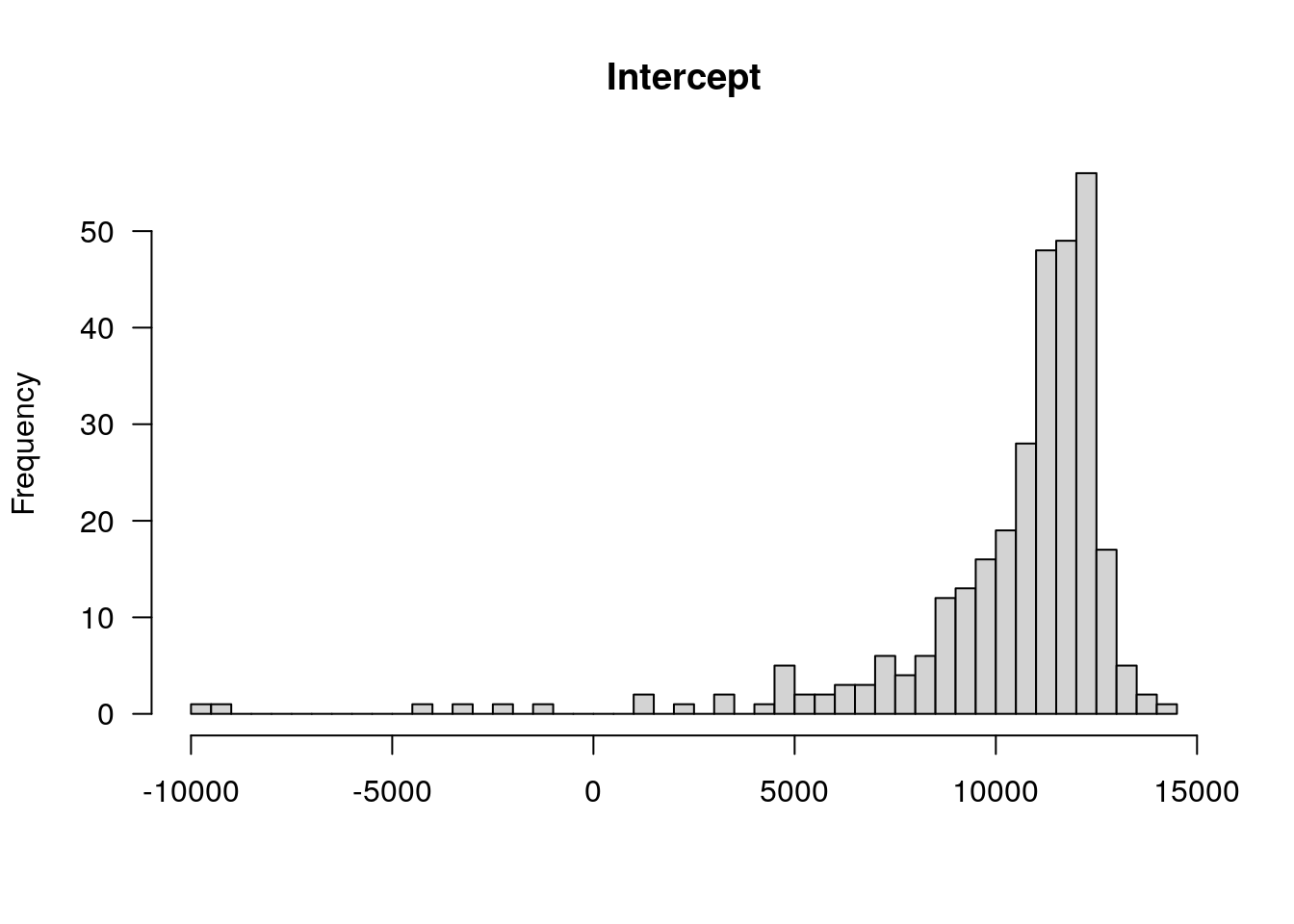
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
plotPheno(WT144, pheno.col=13, xlab = "")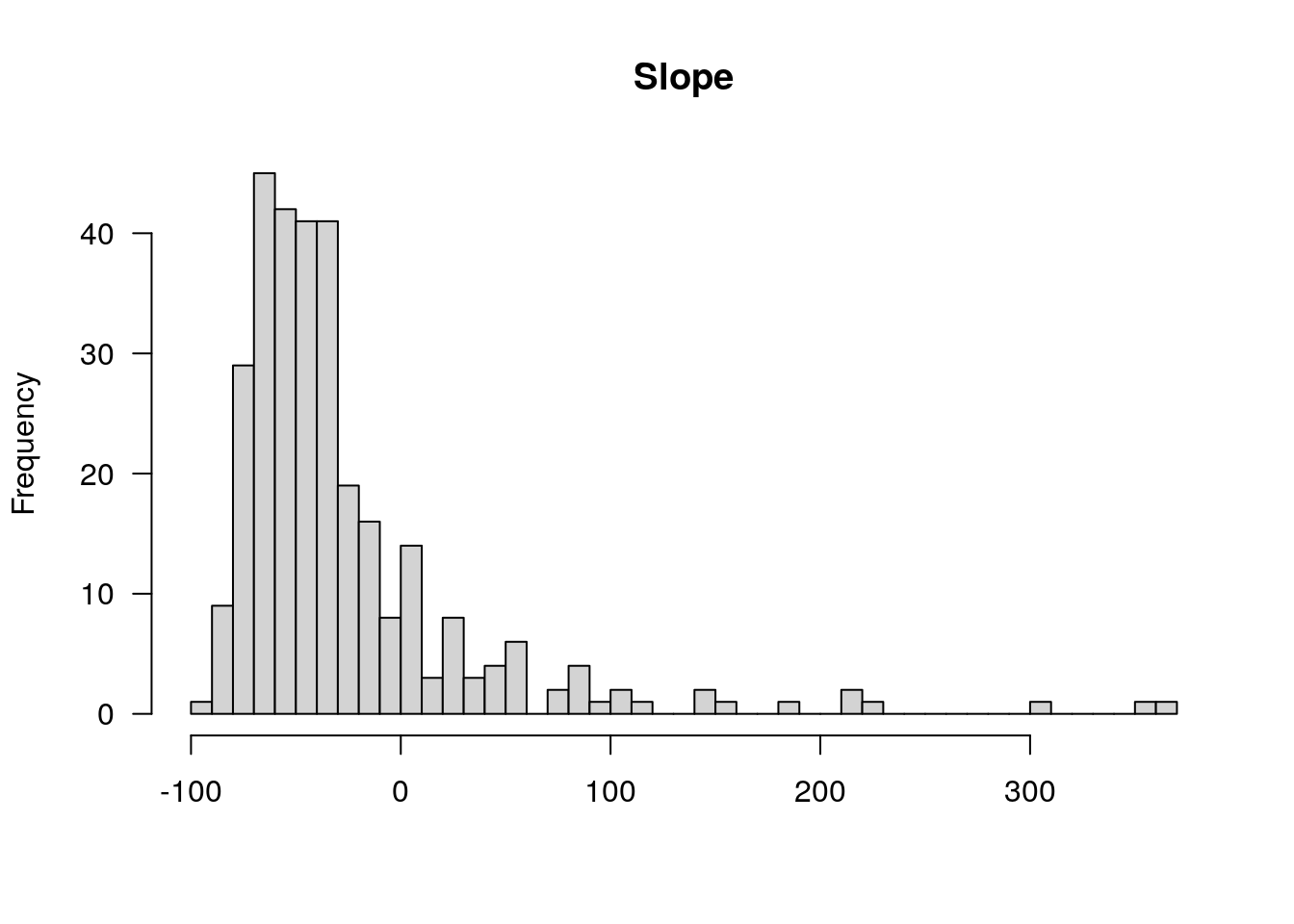
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
Run qtl
#run qtl
print(summary(WT144)) F2 intercross
No. individuals: 309
No. phenotypes: 13
Percent phenotyped: 100 100 100 99.7 100 100 99.7 90.9 100 99.7 90.9 100 100
No. chromosomes: 18
Autosomes: 1 2 3 4 5 7 8 9 10 11 12 13 14 15 16 17 18 19
Total markers: 78
No. markers: 5 8 7 1 22 4 3 5 1 1 4 2 4 2 2 2 2 3
Percent genotyped: 98
Genotypes (%): AA:25.9 AB:50.7 BB:23.3 not BB:0.0 not AA:0.0 WT144 <- drop.nullmarkers(WT144)
covars <- model.matrix(~ Gender, WT144$pheno)[,-1]
#idx
idx <- c(11, 13)
out.mr <- operm <- st <- list()
#loop for each pheno
for (i in 1:length(idx)){
out.mr[[i]] <- scanone(WT144, pheno.col=idx[[i]], method="mr", addcovar=covars)
out.mr[[i]]$lod[is.infinite(out.mr[[i]]$lod)]=0
print(summary(out.mr[[i]]))
#operm
operm[[i]] <-scanone(WT144, pheno.col=idx[[i]], n.perm=1000, method="mr", addcovar=covars)
print(summary(operm[[i]]))
#scantwo
st[[i]] <- scantwo(WT144, pheno.col = idx[[i]], method="mr")
}Warning in checkcovar(cross, pheno.col, addcovar, intcovar, perm.strata, : Dropping 28 individuals with missing phenotypes. chr pos lod
chr1-188155143-.-G-A 1 92.2 1.1232
chr2-137498061-.-T-A 2 67.9 1.8838
chr3-98881230-.-G-A 3 42.9 1.1117
chr4-32821418-.-C-T 4 14.4 0.8050
chr5-136844385-.-T-G 5 76.1 26.1801
chr7-35061547-.-A-G 7 20.9 0.4206
chr8-112465004-.-G-A 8 58.6 0.1365
chr9-64698947-.-C-T 9 34.9 0.2296
chr10-24134777-.-A-T 10 11.5 2.0508
chr11-96730656-.-A-G 11 60.1 0.0908
chr12-43136399-.-A-T 12 19.5 0.9613
chr13-115031098-.-T-C 13 64.6 2.2455
chr14-69691477-.-G-T 14 36.1 6.0905
chr15-58605950-.-C-G 15 25.0 0.0869
chr16-32593683-.-A-G 16 23.0 0.2049
chr17-69219676-.-G-A 17 40.3 0.7094
chr18-51300216-.-T-A 18 27.9 0.1625
chr19-45958839-.-A-T 19 38.8 0.7538Warning in checkcovar(cross, pheno.col, addcovar, intcovar, perm.strata, : Dropping 28 individuals with missing phenotypes.Permutation 20
Permutation 40
Permutation 60
Permutation 80
Permutation 100
Permutation 120
Permutation 140
Permutation 160
Permutation 180
Permutation 200
Permutation 220
Permutation 240
Permutation 260
Permutation 280
Permutation 300
Permutation 320
Permutation 340
Permutation 360
Permutation 380
Permutation 400
Permutation 420
Permutation 440
Permutation 460
Permutation 480
Permutation 500
Permutation 520
Permutation 540
Permutation 560
Permutation 580
Permutation 600
Permutation 620
Permutation 640
Permutation 660
Permutation 680
Permutation 700
Permutation 720
Permutation 740
Permutation 760
Permutation 780
Permutation 800
Permutation 820
Permutation 840
Permutation 860
Permutation 880
Permutation 900
Permutation 920
Permutation 940
Permutation 960
Permutation 980
Permutation 1000
LOD thresholds (1000 permutations)
lod
5% 3.10
10% 2.68Warning in checkcovar(cross, pheno.col, addcovar, intcovar, perm.strata, : Dropping 28 individuals with missing phenotypes. --Running scanone
--Running scantwo
(1,1)
(1,2)
(1,3)
(1,4)
(1,5)
(1,7)
(1,8)
(1,9)
(1,10)
(1,11)
(1,12)
(1,13)
(1,14)
(1,15)
(1,16)
(1,17)
(1,18)
(1,19)
(2,2)
(2,3)
(2,4)
(2,5)
(2,7)
(2,8)
(2,9)
(2,10)
(2,11)
(2,12)
(2,13)
(2,14)
(2,15)
(2,16)
(2,17)
(2,18)
(2,19)
(3,3)
(3,4)
(3,5)
(3,7)
(3,8)
(3,9)
(3,10)
(3,11)
(3,12)
(3,13)
(3,14)
(3,15)
(3,16)
(3,17)
(3,18)
(3,19)
(4,4)
(4,5)
(4,7)
(4,8)
(4,9)
(4,10)
(4,11)
(4,12)
(4,13)
(4,14)
(4,15)
(4,16)
(4,17)
(4,18)
(4,19)
(5,5)
(5,7)
(5,8)
(5,9)
(5,10)
(5,11)
(5,12)
(5,13)
(5,14)
(5,15)
(5,16)
(5,17)
(5,18)
(5,19)
(7,7)
(7,8)
(7,9)
(7,10)
(7,11)
(7,12)
(7,13)
(7,14)
(7,15)
(7,16)
(7,17)
(7,18)
(7,19)
(8,8)
(8,9)
(8,10)
(8,11)
(8,12)
(8,13)
(8,14)
(8,15)
(8,16)
(8,17)
(8,18)
(8,19)
(9,9)
(9,10)
(9,11)
(9,12)
(9,13)
(9,14)
(9,15)
(9,16)
(9,17)
(9,18)
(9,19)
(10,10)
(10,11)
(10,12)
(10,13)
(10,14)
(10,15)
(10,16)
(10,17)
(10,18)
(10,19)
(11,11)
(11,12)
(11,13)
(11,14)
(11,15)
(11,16)
(11,17)
(11,18)
(11,19)
(12,12)
(12,13)
(12,14)
(12,15)
(12,16)
(12,17)
(12,18)
(12,19)
(13,13)
(13,14)
(13,15)
(13,16)
(13,17)
(13,18)
(13,19)
(14,14)
(14,15)
(14,16)
(14,17)
(14,18)
(14,19)
(15,15)
(15,16)
(15,17)
(15,18)
(15,19)
(16,16)
(16,17)
(16,18)
(16,19)
(17,17)
(17,18)
(17,19)
(18,18)
(18,19)
(19,19)
chr pos lod
chr1-65643621-.-C-A 1 33.3 1.4142
chr2-137498061-.-T-A 2 67.9 2.3977
chr3-59218564-.-T-A 3 29.0 1.5114
chr4-32821418-.-C-T 4 14.4 0.9584
chr5-136332619-.-C-A 5 76.0 28.2723
chr7-35061547-.-A-G 7 20.9 0.2899
chr8-112465004-.-G-A 8 58.6 0.2728
chr9-69415567-.-T-A 9 38.6 0.5506
chr10-24134777-.-A-T 10 11.5 2.1472
chr11-96730656-.-A-G 11 60.1 0.2845
chr12-43136399-.-A-T 12 19.5 1.1682
chr13-115031098-.-T-C 13 64.6 2.0179
chr14-69691477-.-G-T 14 36.1 6.3651
chr15-58605950-.-C-G 15 25.0 0.2664
chr16-32593683-.-A-G 16 23.0 0.4304
chr17-69219676-.-G-A 17 40.3 0.9353
chr18-89347666-.-A-G 18 59.0 0.0746
chr19-45958839-.-A-T 19 38.8 0.5790
Permutation 20
Permutation 40
Permutation 60
Permutation 80
Permutation 100
Permutation 120
Permutation 140
Permutation 160
Permutation 180
Permutation 200
Permutation 220
Permutation 240
Permutation 260
Permutation 280
Permutation 300
Permutation 320
Permutation 340
Permutation 360
Permutation 380
Permutation 400
Permutation 420
Permutation 440
Permutation 460
Permutation 480
Permutation 500
Permutation 520
Permutation 540
Permutation 560
Permutation 580
Permutation 600
Permutation 620
Permutation 640
Permutation 660
Permutation 680
Permutation 700
Permutation 720
Permutation 740
Permutation 760
Permutation 780
Permutation 800
Permutation 820
Permutation 840
Permutation 860
Permutation 880
Permutation 900
Permutation 920
Permutation 940
Permutation 960
Permutation 980
Permutation 1000
LOD thresholds (1000 permutations)
lod
5% 3.10
10% 2.75
--Running scanone
--Running scantwo
(1,1)
(1,2)
(1,3)
(1,4)
(1,5)
(1,7)
(1,8)
(1,9)
(1,10)
(1,11)
(1,12)
(1,13)
(1,14)
(1,15)
(1,16)
(1,17)
(1,18)
(1,19)
(2,2)
(2,3)
(2,4)
(2,5)
(2,7)
(2,8)
(2,9)
(2,10)
(2,11)
(2,12)
(2,13)
(2,14)
(2,15)
(2,16)
(2,17)
(2,18)
(2,19)
(3,3)
(3,4)
(3,5)
(3,7)
(3,8)
(3,9)
(3,10)
(3,11)
(3,12)
(3,13)
(3,14)
(3,15)
(3,16)
(3,17)
(3,18)
(3,19)
(4,4)
(4,5)
(4,7)
(4,8)
(4,9)
(4,10)
(4,11)
(4,12)
(4,13)
(4,14)
(4,15)
(4,16)
(4,17)
(4,18)
(4,19)
(5,5)
(5,7)
(5,8)
(5,9)
(5,10)
(5,11)
(5,12)
(5,13)
(5,14)
(5,15)
(5,16)
(5,17)
(5,18)
(5,19)
(7,7)
(7,8)
(7,9)
(7,10)
(7,11)
(7,12)
(7,13)
(7,14)
(7,15)
(7,16)
(7,17)
(7,18)
(7,19)
(8,8)
(8,9)
(8,10)
(8,11)
(8,12)
(8,13)
(8,14)
(8,15)
(8,16)
(8,17)
(8,18)
(8,19)
(9,9)
(9,10)
(9,11)
(9,12)
(9,13)
(9,14)
(9,15)
(9,16)
(9,17)
(9,18)
(9,19)
(10,10)
(10,11)
(10,12)
(10,13)
(10,14)
(10,15)
(10,16)
(10,17)
(10,18)
(10,19)
(11,11)
(11,12)
(11,13)
(11,14)
(11,15)
(11,16)
(11,17)
(11,18)
(11,19)
(12,12)
(12,13)
(12,14)
(12,15)
(12,16)
(12,17)
(12,18)
(12,19)
(13,13)
(13,14)
(13,15)
(13,16)
(13,17)
(13,18)
(13,19)
(14,14)
(14,15)
(14,16)
(14,17)
(14,18)
(14,19)
(15,15)
(15,16)
(15,17)
(15,18)
(15,19)
(16,16)
(16,17)
(16,18)
(16,19)
(17,17)
(17,18)
(17,19)
(18,18)
(18,19)
(19,19)save(out.mr, operm, st, file = "output/qtl.out.obj.mixedmodel.final.RData")Plot on run qtl
name = "WT144_plot"
#print
print(summary(WT144)) F2 intercross
No. individuals: 309
No. phenotypes: 13
Percent phenotyped: 100 100 100 99.7 100 100 99.7 90.9 100 99.7 90.9 100 100
No. chromosomes: 18
Autosomes: 1 2 3 4 5 7 8 9 10 11 12 13 14 15 16 17 18 19
Total markers: 78
No. markers: 5 8 7 1 22 4 3 5 1 1 4 2 4 2 2 2 2 3
Percent genotyped: 98
Genotypes (%): AA:25.9 AB:50.7 BB:23.3 not BB:0.0 not AA:0.0 WT144 <- drop.nullmarkers(WT144)
covars <- model.matrix(~ Gender, WT144$pheno)[,-1]
#idx
idx <- c(11, 13)
#map
map <- purrr::map(WT144$geno, ~(.x$map))
attr(map, "is_x_chr") <- structure(c(rep(FALSE,18)), names=c(1:5, 7:19))
load("output/qtl.out.obj.mixedmodel.final.RData")
#loop for each pheno
for (i in 1:length(idx)){
print(colnames(WT144$pheno)[idx[[i]]])
#operm_hist
#pdf(paste0("output/", name,"-", colnames(WT144$pheno)[idx[[i]]], ".pdf"), width = 10, height = 10)
plot(operm[[i]][!is.infinite(operm[[i]])], main=paste(name, colnames(WT144$pheno)[idx[[i]]], sep="-"))
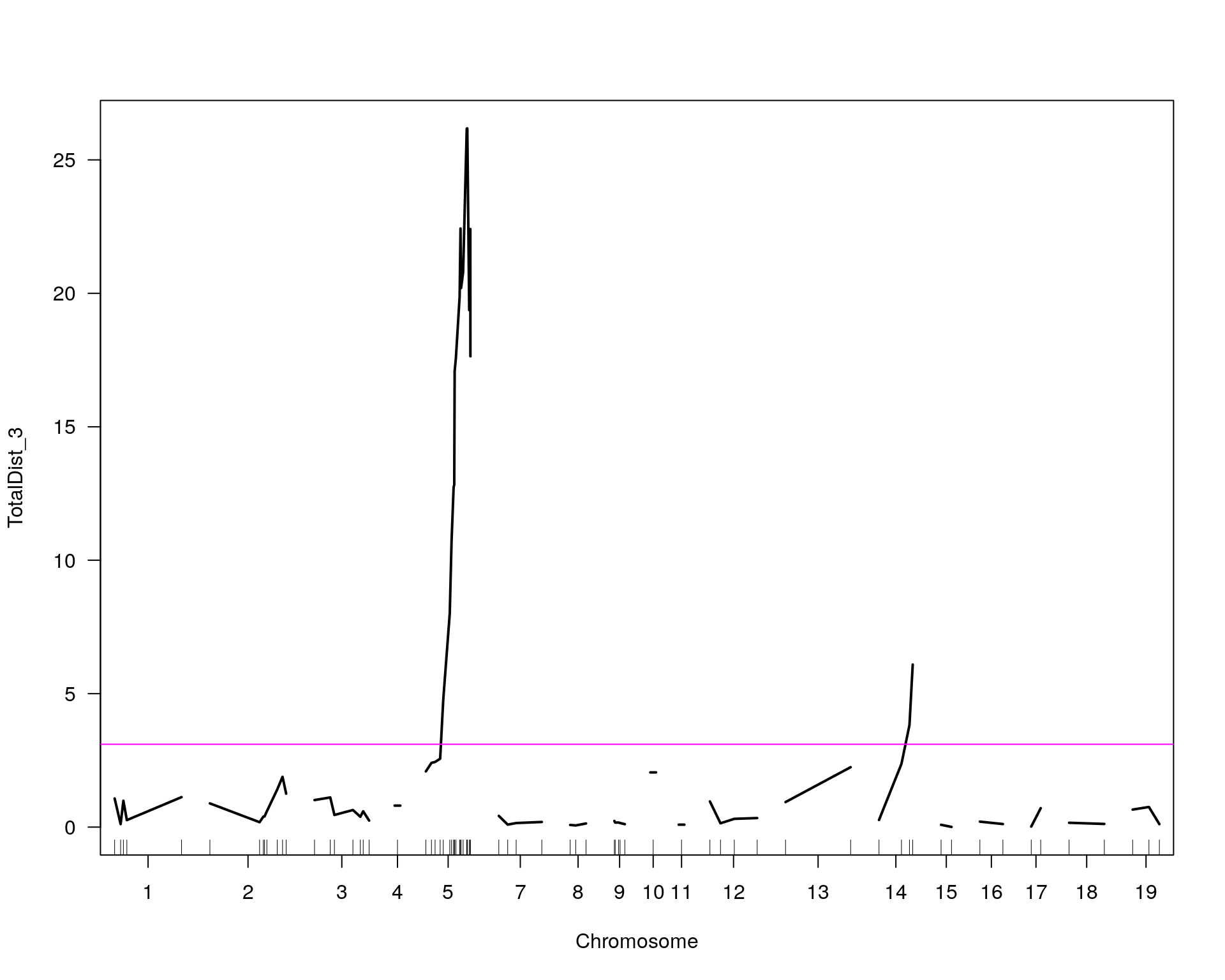
#mrscan_pheno
plot(out.mr[[i]], ylab=colnames(WT144$pheno)[idx[[i]]])
add.threshold(out.mr[[i]], perms = operm[[i]], alpha = 0.05, col="magenta")
#pxg
peak = summary(out.mr[[i]], threshold=summary(operm[[i]], alpha = 0.05)[[1]])
print(peak)
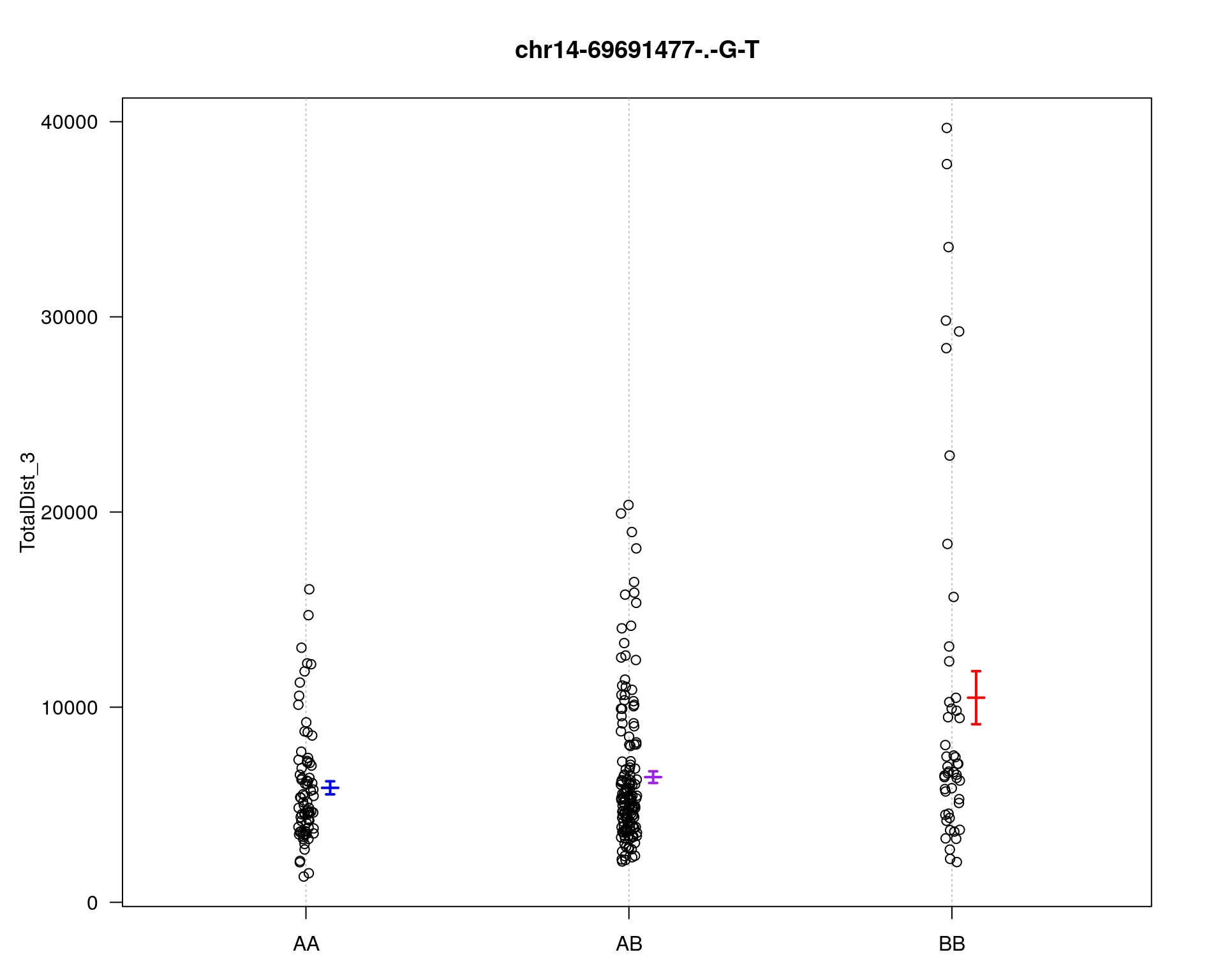
marker = c("chr5-136332619-.-C-A", "chr14-69691477-.-G-T")
for (mar in marker){
par(mar=c(3, 5, 4, 3))
plotPXG(WT144, marker=mar, jitter = 0.25, pheno.col = idx[[i]], infer=F, main=paste(mar),
mgp=c(3.4,1,0))
}
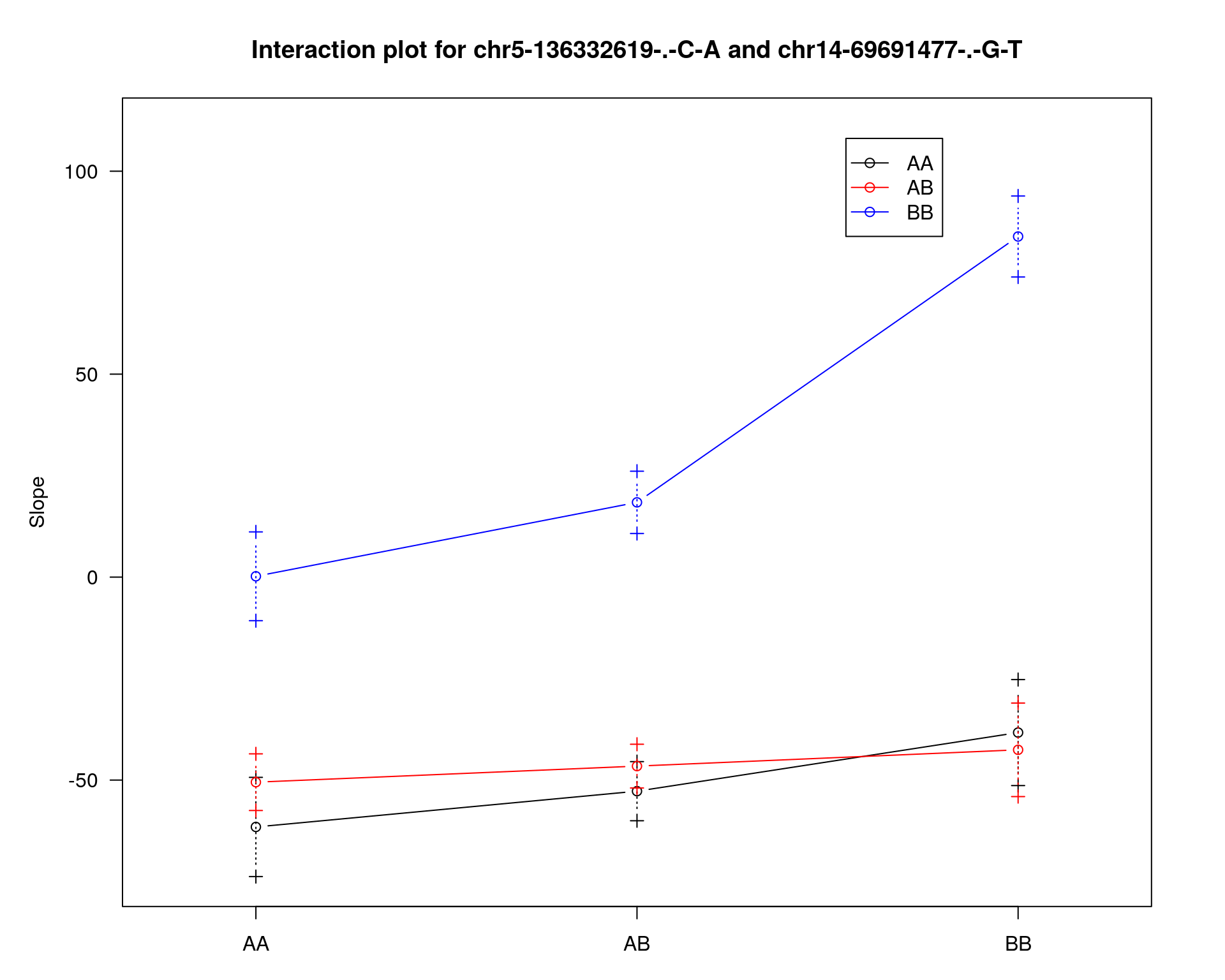
#interaction_effect
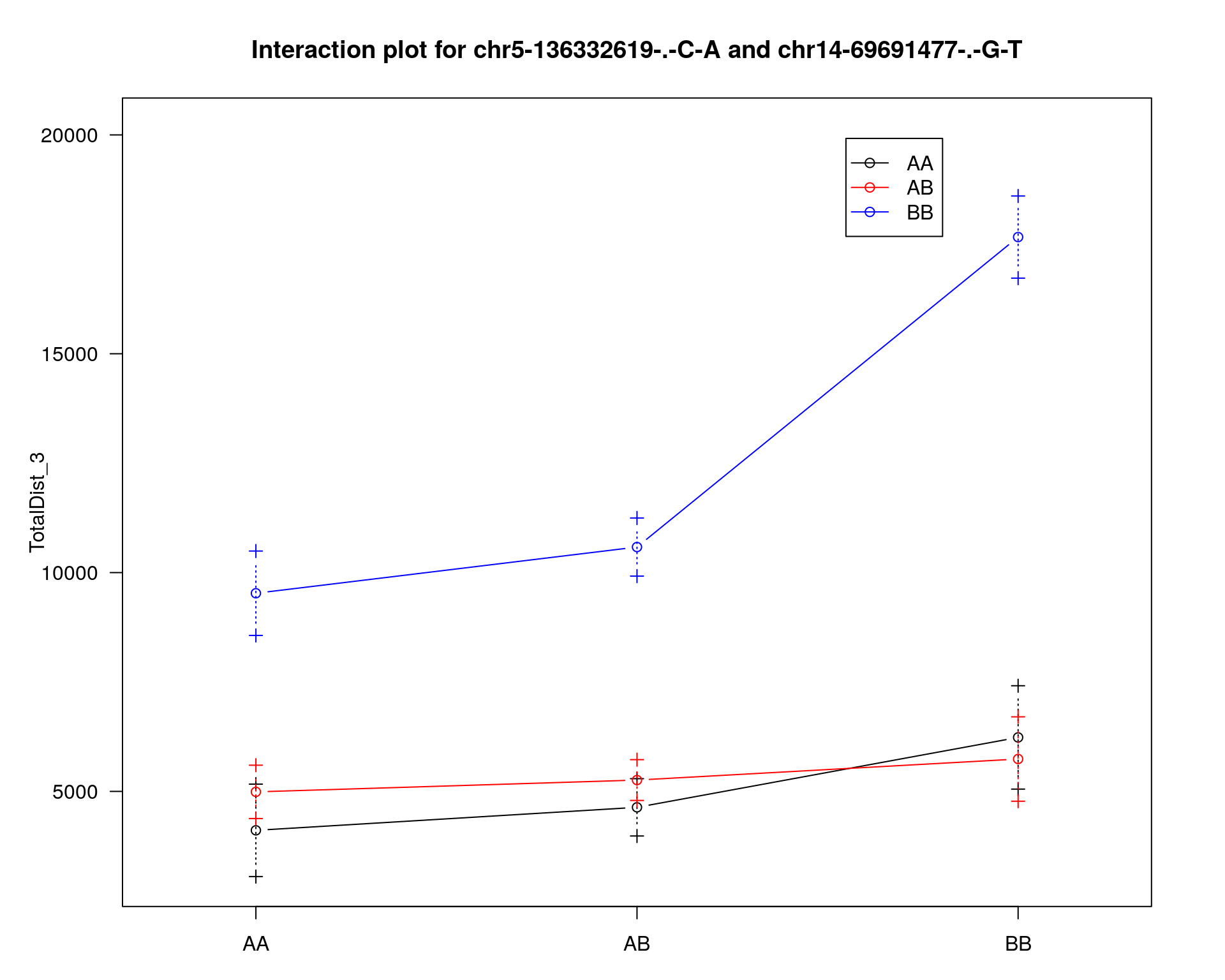
effectplot(WT144,
mname1 = marker[[1]],
mname2 = marker[[2]],
pheno.col = idx[[i]], add.legend=T, legend.lab = "")
##Multiple-QTL analyses
# After performing the single- and two-QTL genome scans, it’s best to bring the identified loci together into a joint model, which we then refine and from which we may explore the possibility of further QTL. In this effort, we work with “QTL objects” created by makeqtl(). We fit multiple-QTL models with fitqtl(). A number of additional functions will be introduced below.
#First, we create a QTL object containing the loci on chr 5 and 14.
#chr5-136332619-.-C-A 75.97770
#chr14-69691477-.-G-T 36.07219
WT144 <- sim.geno(WT144, n.draws=64)
qtl <- makeqtl(WT144, chr=c(5,14), pos=c(75.97770, 36.07219), what="draws")
out.fq <- fitqtl(WT144, pheno.col=idx[[i]], qtl=qtl)
print(summary(out.fq))
#We may obtain the estimated effects of the QTL via get.ests=TRUE. We use dropone=FALSE to suppress the drop-one-term analysis.
print(summary(fitqtl(WT144, pheno.col=idx[[i]], qtl=qtl, get.ests=TRUE, dropone=FALSE)))
#To assess the possibility of an interaction between the two QTL, we may fit the model with the interaction, indicated via a model “formula”.
print("To assess the possibility of an interaction between the two QTL")
out.fqi <- fitqtl(WT144, pheno.col=idx[[i]], qtl=qtl, formula=y~Q1+Q2+Q1:Q2)
print(summary(out.fqi))
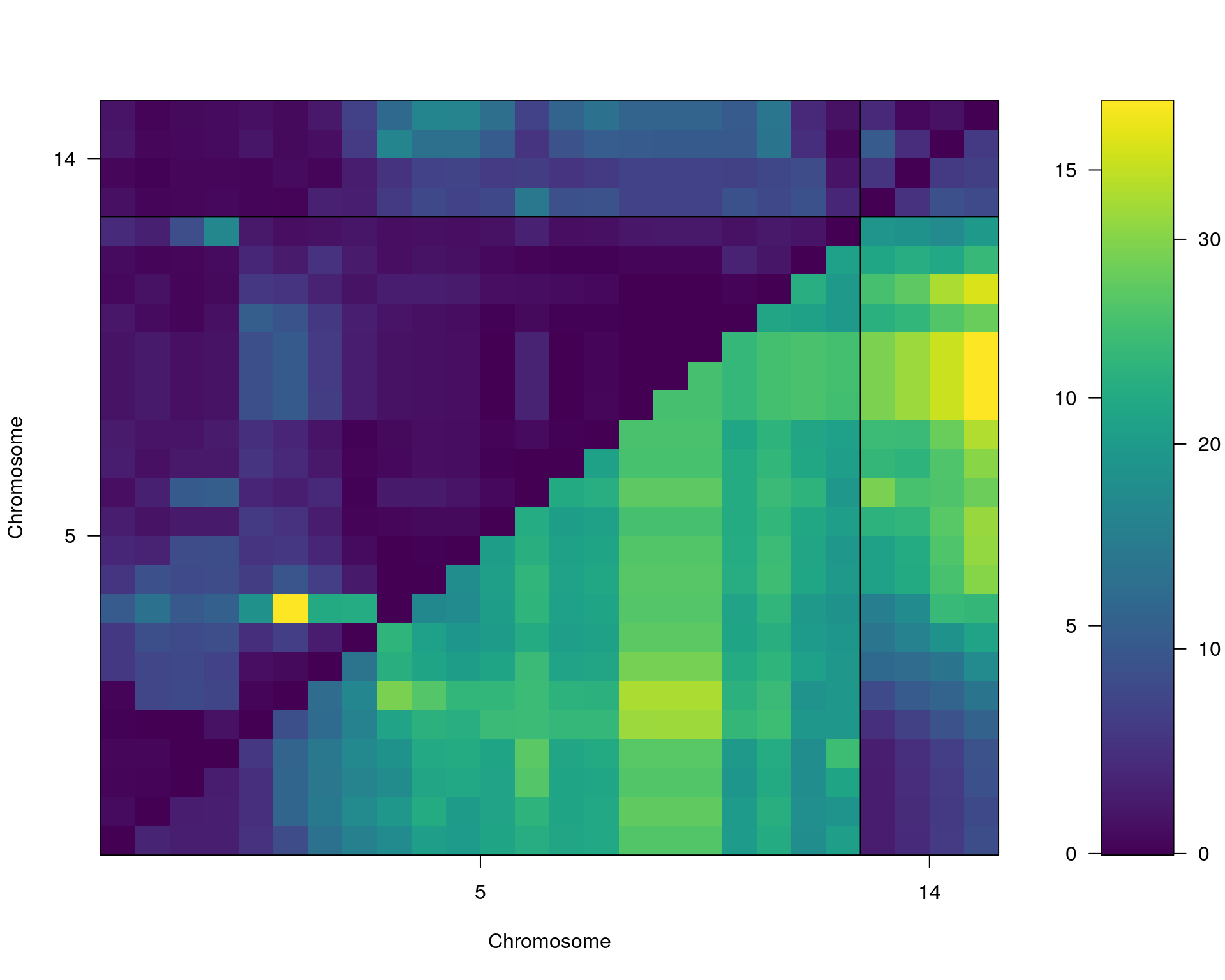
#complete_scan2
plot(st[[i]])
#toptwo_scan2
plot(st[[i]], chr = summary(out.mr[[i]])[order(-summary(out.mr[[i]])$lod), "chr"][1:2])
#dev.off()
}[1] "TotalDist_3"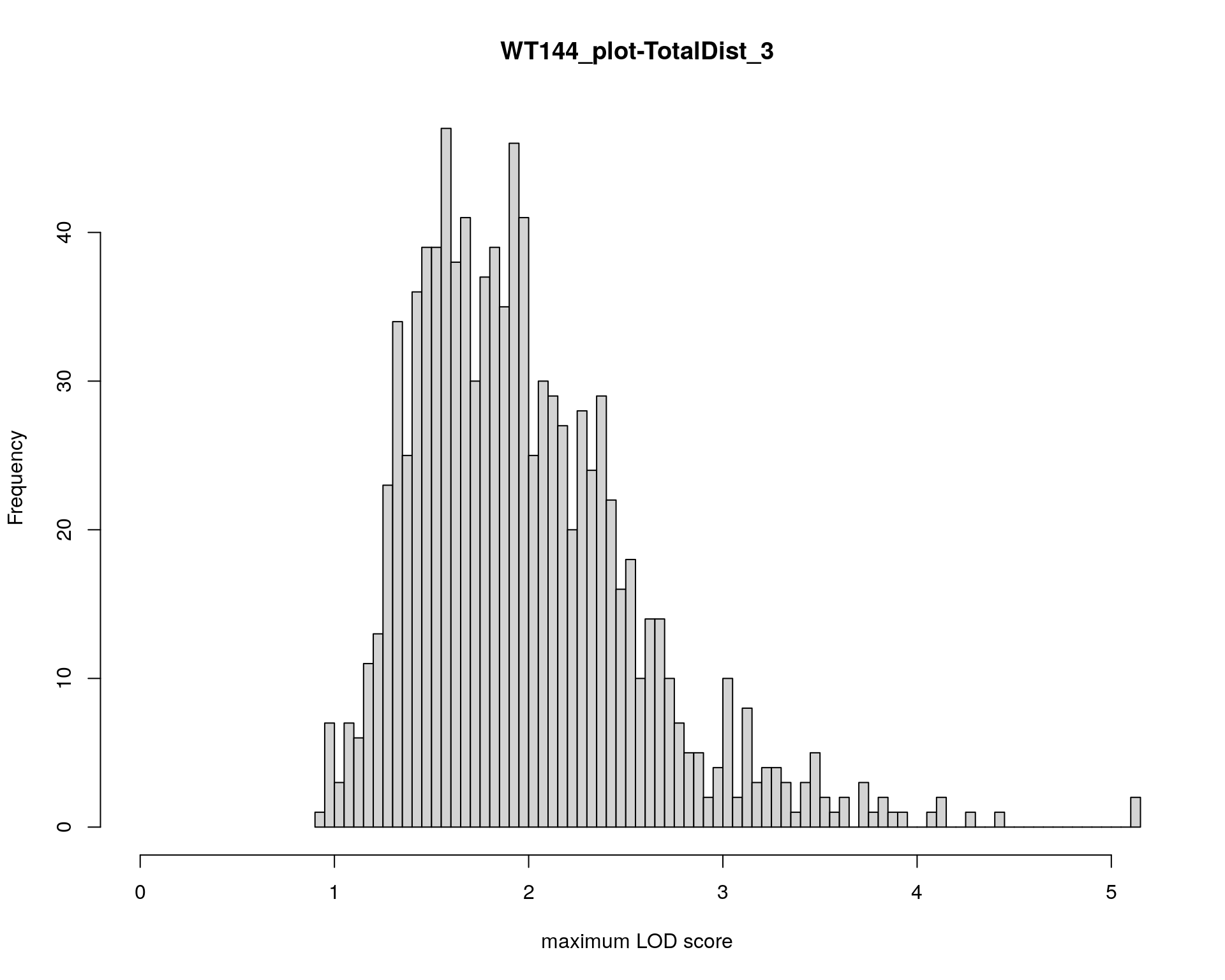
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
chr pos lod
chr5-136332619-.-C-A 5 76.0 26.18
chr14-69691477-.-G-T 14 36.1 6.09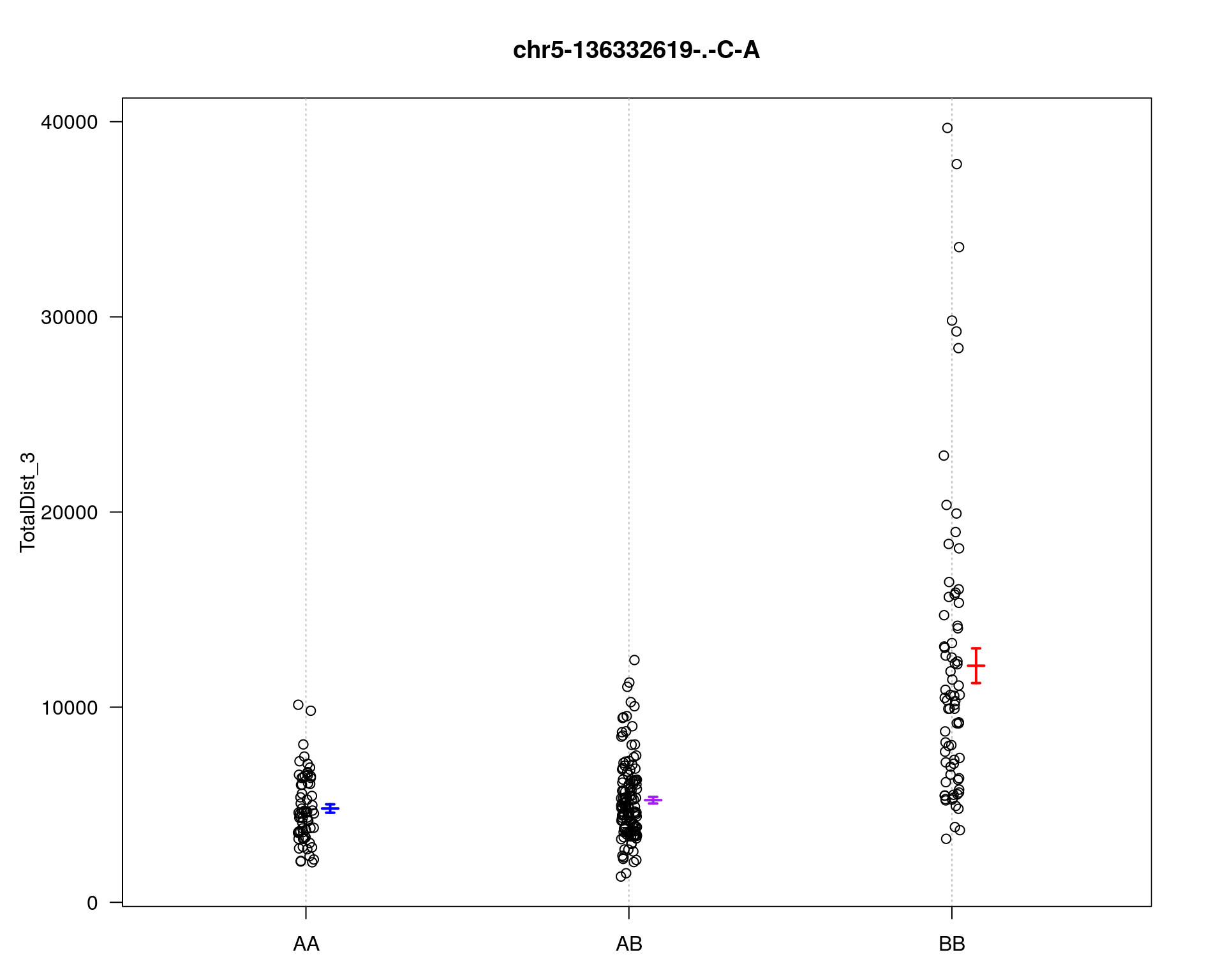
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 281
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 3170953192 792738298 31.45606 40.28087 0 0
Error 276 4701152932 17033163
Total 280 7872106124
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 2 2.423e+09 25.367 30.783 71.13 0 < 2e-16 ***
14@36.1 2 4.478e+08 5.552 5.688 13.14 0 3.52e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 281
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 3170953192 792738298 31.45606 40.28087 0 0
Error 276 4701152932 17033163
Total 280 7872106124
Estimated effects:
-----------------
est SE t
Intercept 7106.1 251.6 28.248
5@76.0a 3549.0 349.2 10.162
5@76.0d -2908.4 497.5 -5.846
14@36.1a 1818.7 380.1 4.785
14@36.1d -1335.8 503.4 -2.654
[1] "To assess the possibility of an interaction between the two QTL"
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 281
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q1:Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 8 3558421585 444802698 36.70458 45.20292 0 0
Error 272 4313684539 15859134
Total 280 7872106124
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 6 2.811e+09 30.615 35.705 29.539 0 < 2e-16 ***
14@36.1 6 8.353e+08 10.800 10.610 8.778 0 9.42e-09 ***
5@76.0:14@36.1 4 3.875e+08 5.249 4.922 6.108 0 0.000101 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1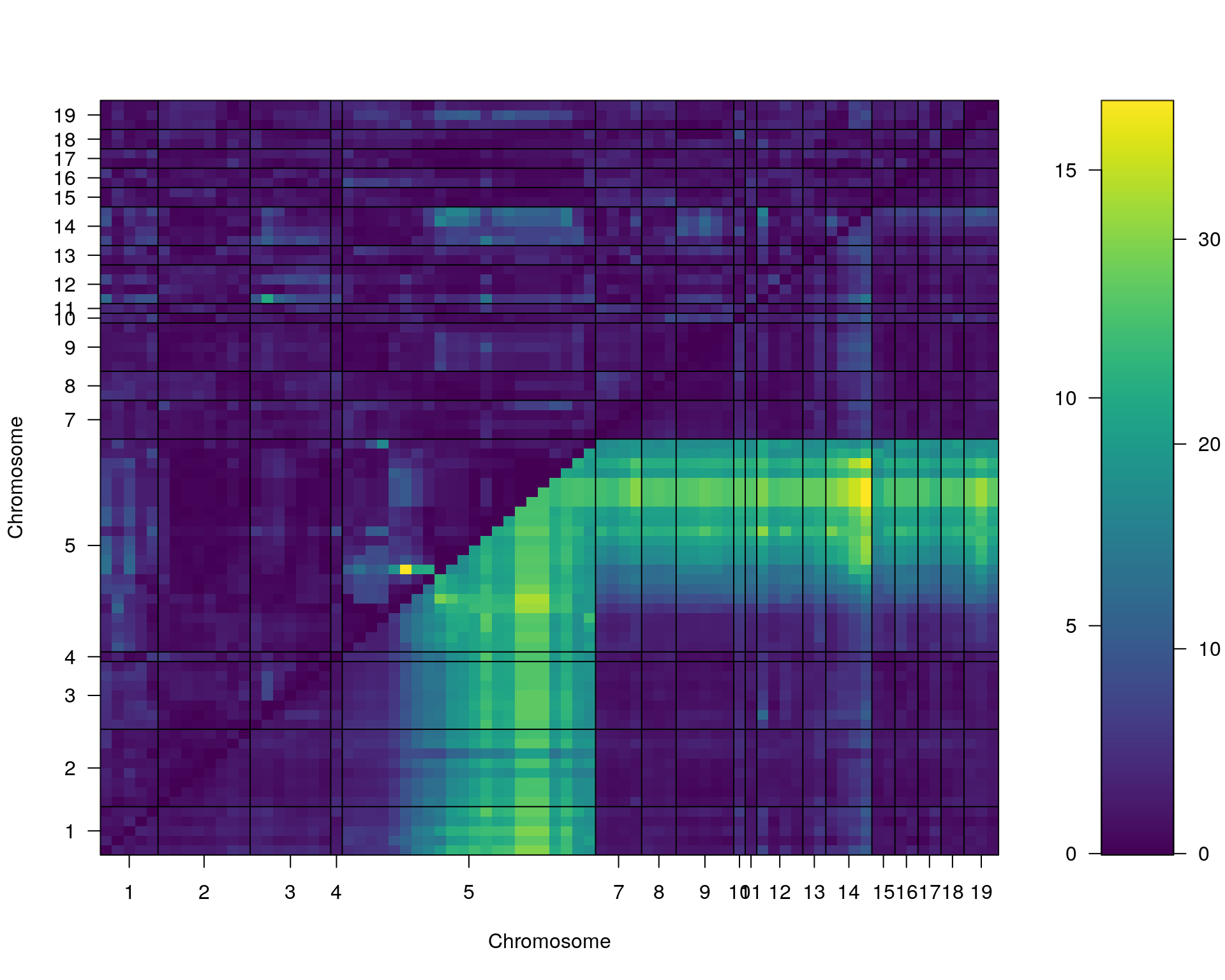
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
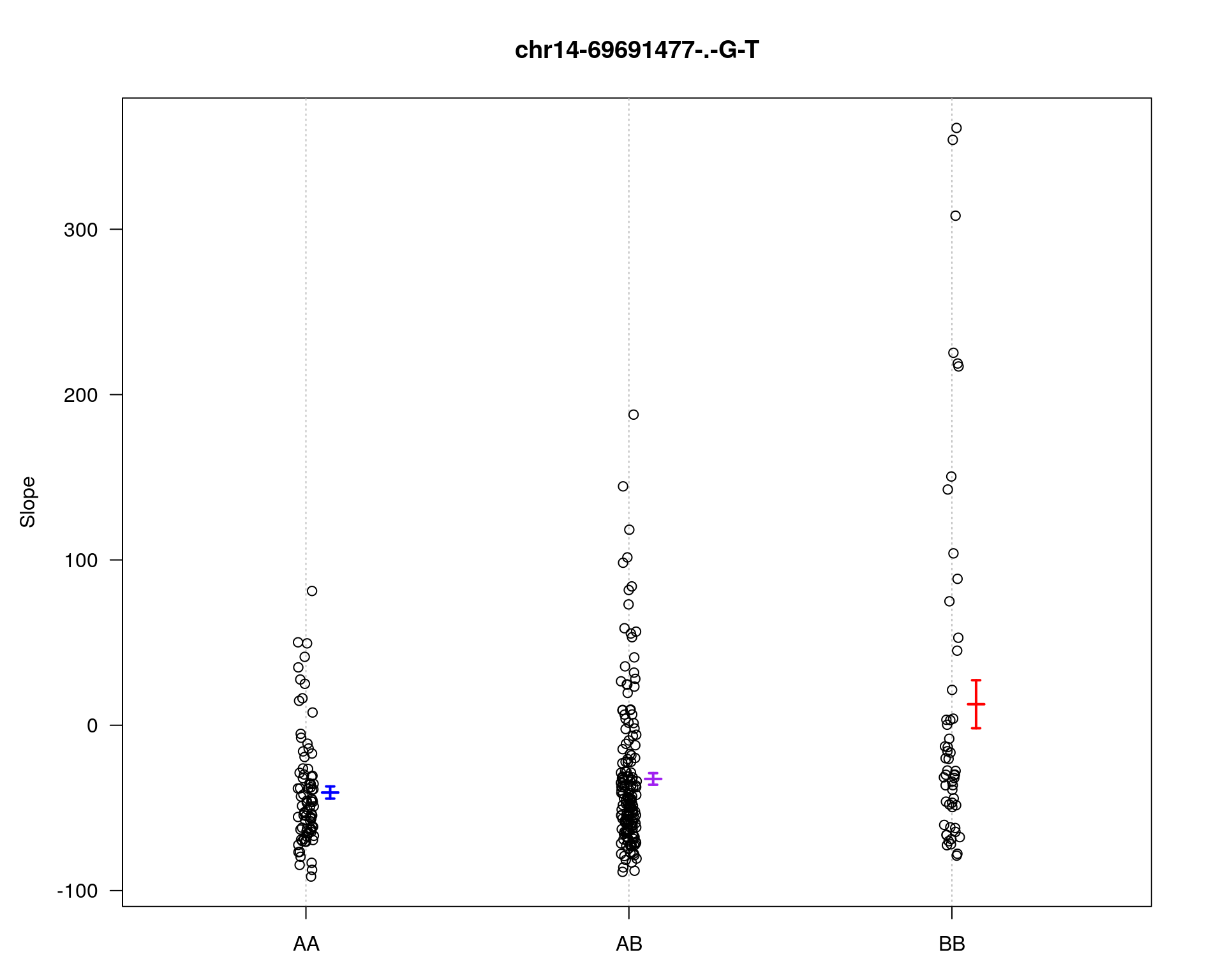
[1] "Slope"
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
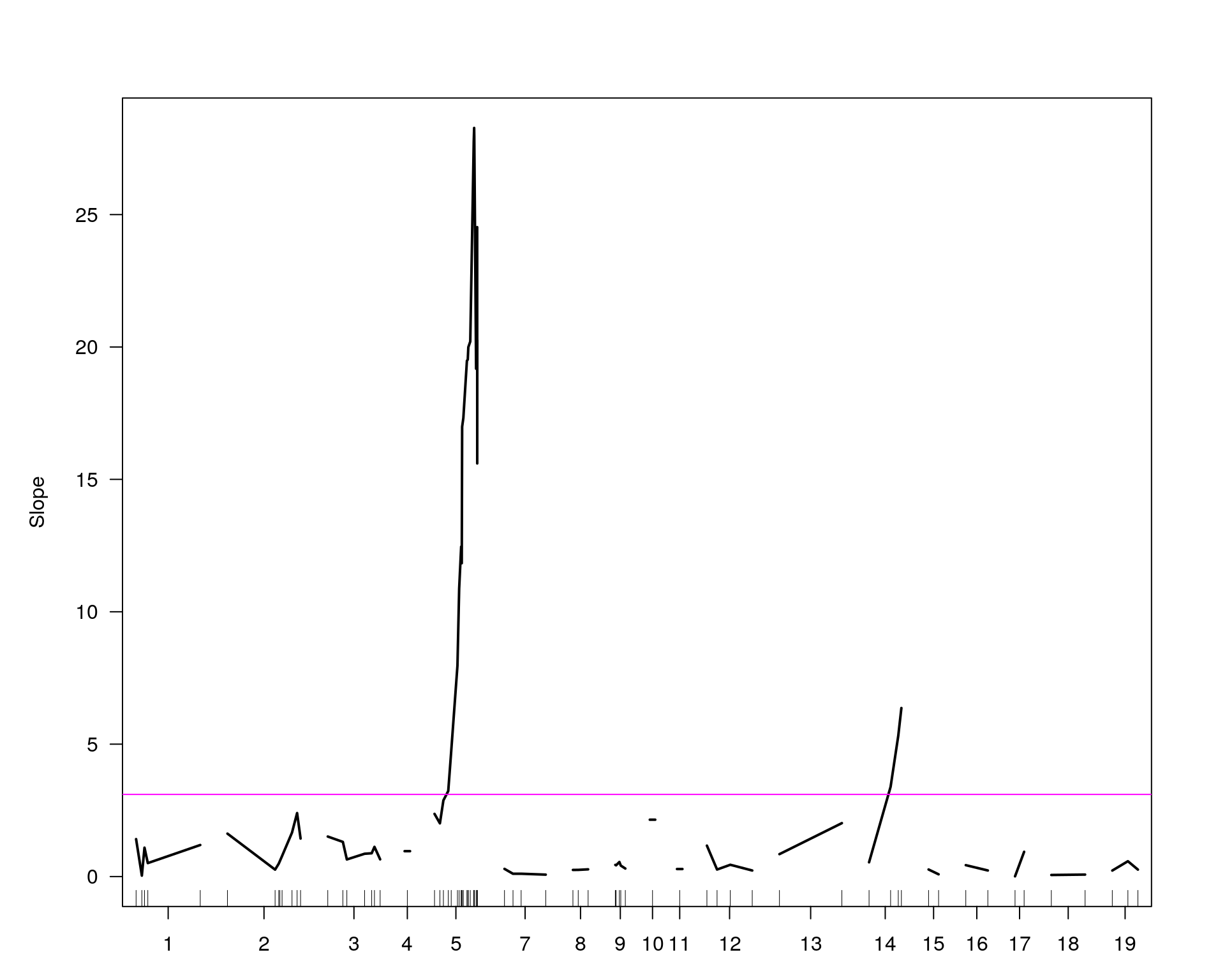
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
chr pos lod
chr5-136332619-.-C-A 5 76.0 28.27
chr14-69691477-.-G-T 14 36.1 6.37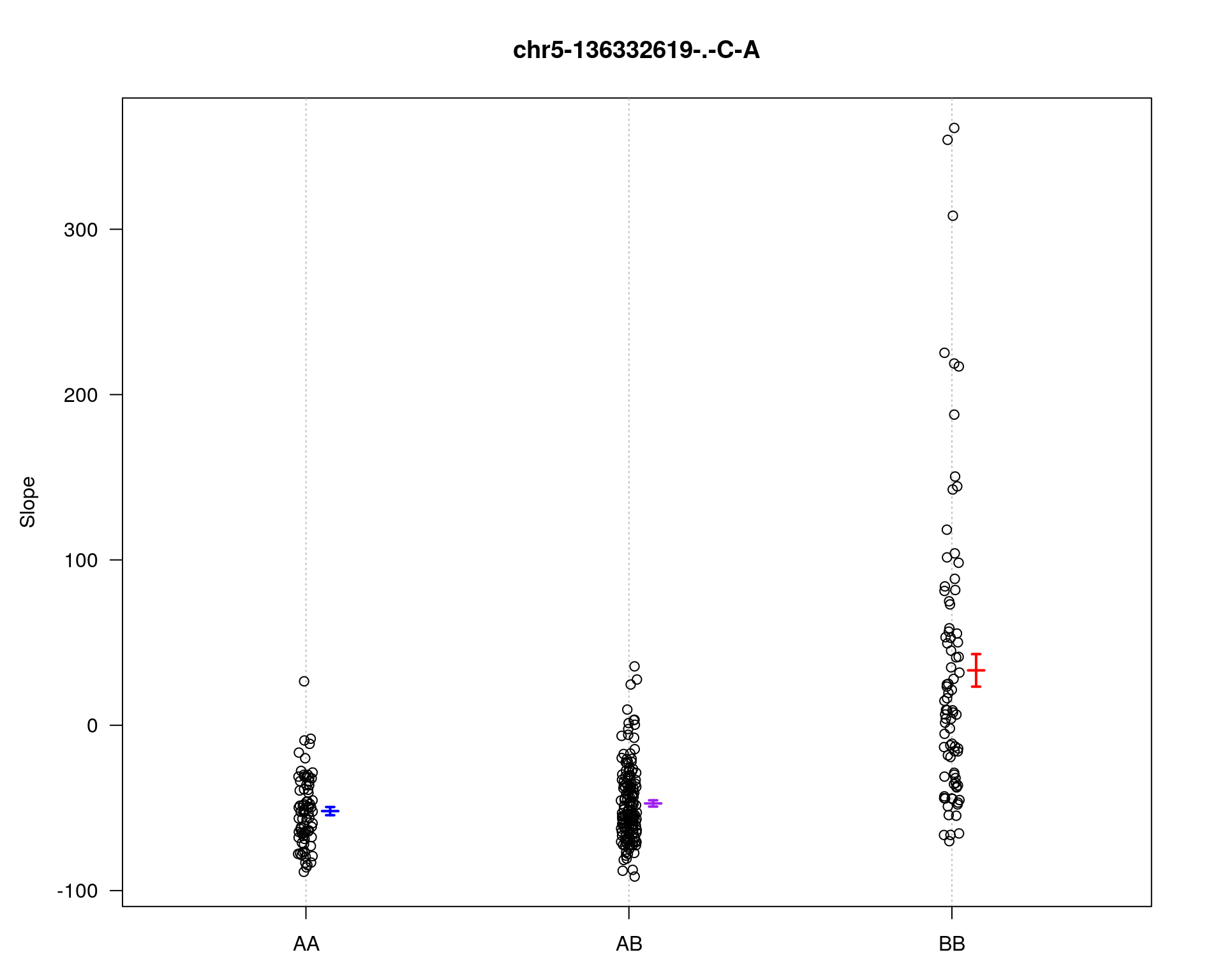
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 309
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 464953.1 116238.280 32.79348 38.6599 0 0
Error 304 737722.4 2426.718
Total 308 1202675.5
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 2 355772 26.408 29.582 73.30 0 < 2e-16 ***
14@36.1 2 55117 4.835 4.583 11.36 0 1.75e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 309
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 464953.1 116238.280 32.79348 38.6599 0 0
Error 304 737722.4 2426.718
Total 308 1202675.5
Estimated effects:
-----------------
est SE t
Intercept -26.034 2.858 -9.109
5@76.0a 40.855 3.927 10.405
5@76.0d -33.307 5.693 -5.850
14@36.1a 19.763 4.307 4.589
14@36.1d -11.513 5.726 -2.011
[1] "To assess the possibility of an interaction between the two QTL"
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 309
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q1:Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 8 507503.1 63437.888 36.77964 42.19784 0 0
Error 300 695172.4 2317.241
Total 308 1202675.5
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 6 398322 30.394 33.120 28.649 0.000 < 2e-16 ***
14@36.1 6 97667 8.821 8.121 7.025 0.000 5.22e-07 ***
5@76.0:14@36.1 4 42550 3.986 3.538 4.591 0.001 0.0013 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1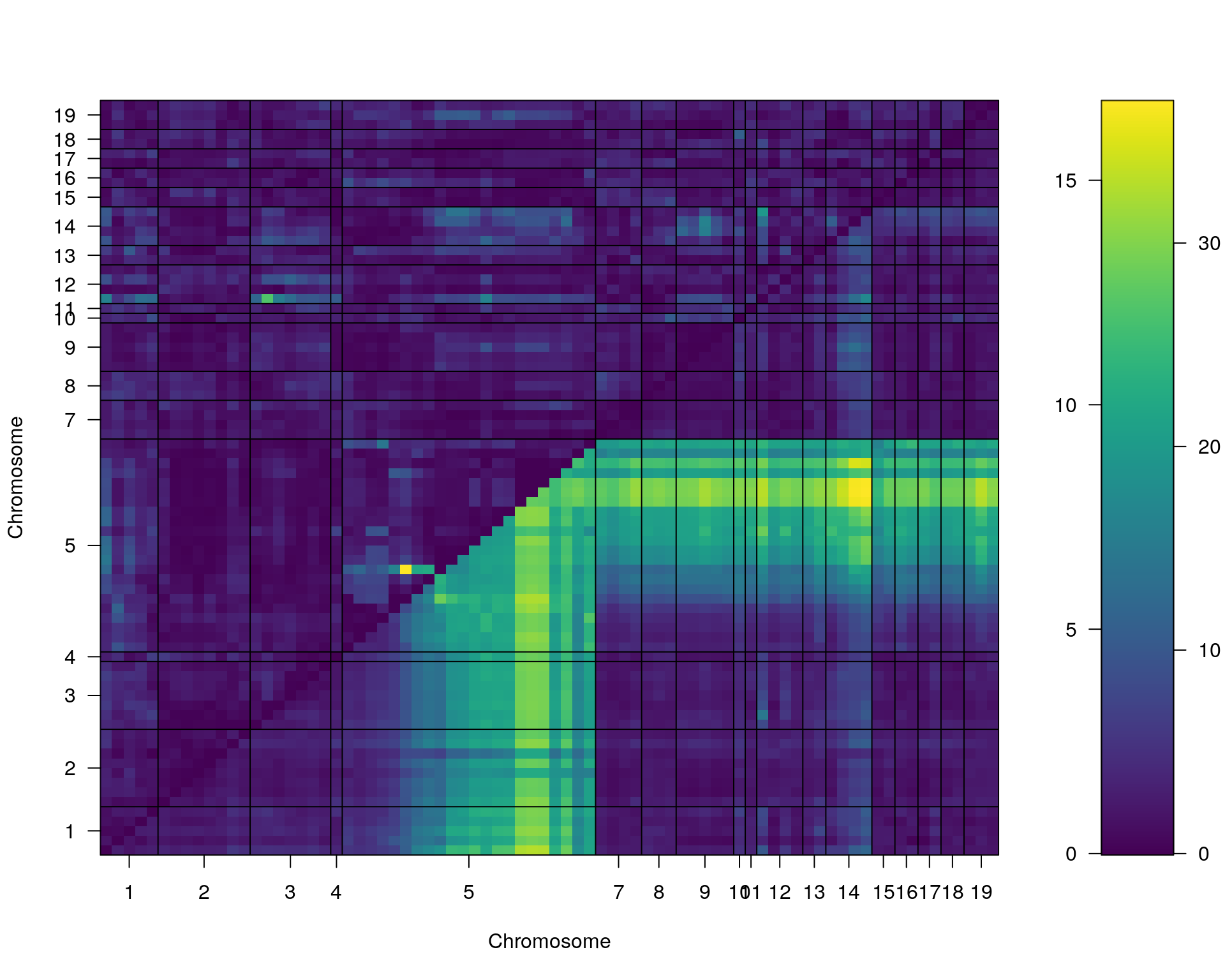
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#plot phenotypes: slope and totaldistance3
plot(out.mr[[2]], col= "blue", ylim = c(0, 30), ylab = "LOD score")
plot(out.mr[[1]], col= "green", add=TRUE)
abline(h=3.2, col="red", lty=2, lwd=3)
# Add a legend
legend("topright",
legend=c("Slope", "Total distance traveled"),
col=c("blue", "green"), lty=1, cex=0.8)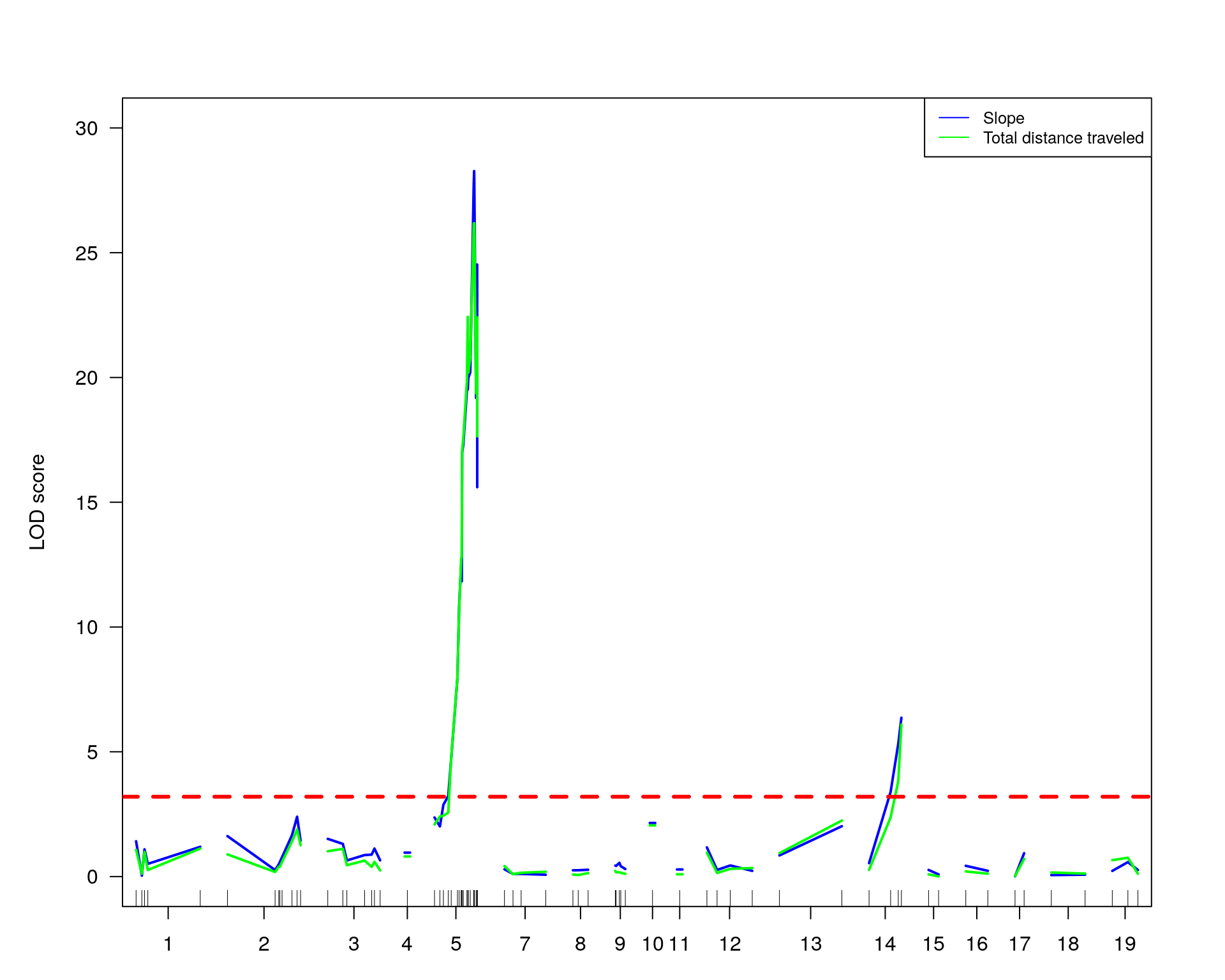
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#only at chr 5 and 14
#plot phenotypes: slope and totaldistance3
plot(out.mr[[2]], col= "blue", ylim = c(0, 30), ylab = "LOD score", chr = c(5, 14))
plot(out.mr[[1]], col= "green", add=TRUE, chr = c(5, 14))
abline(h=3.2, col="red", lty=2, lwd=3)
# Add a legend
legend("topright",
legend=c("Slope", "Total distance traveled"),
col=c("blue", "green"), lty=1, cex=0.8)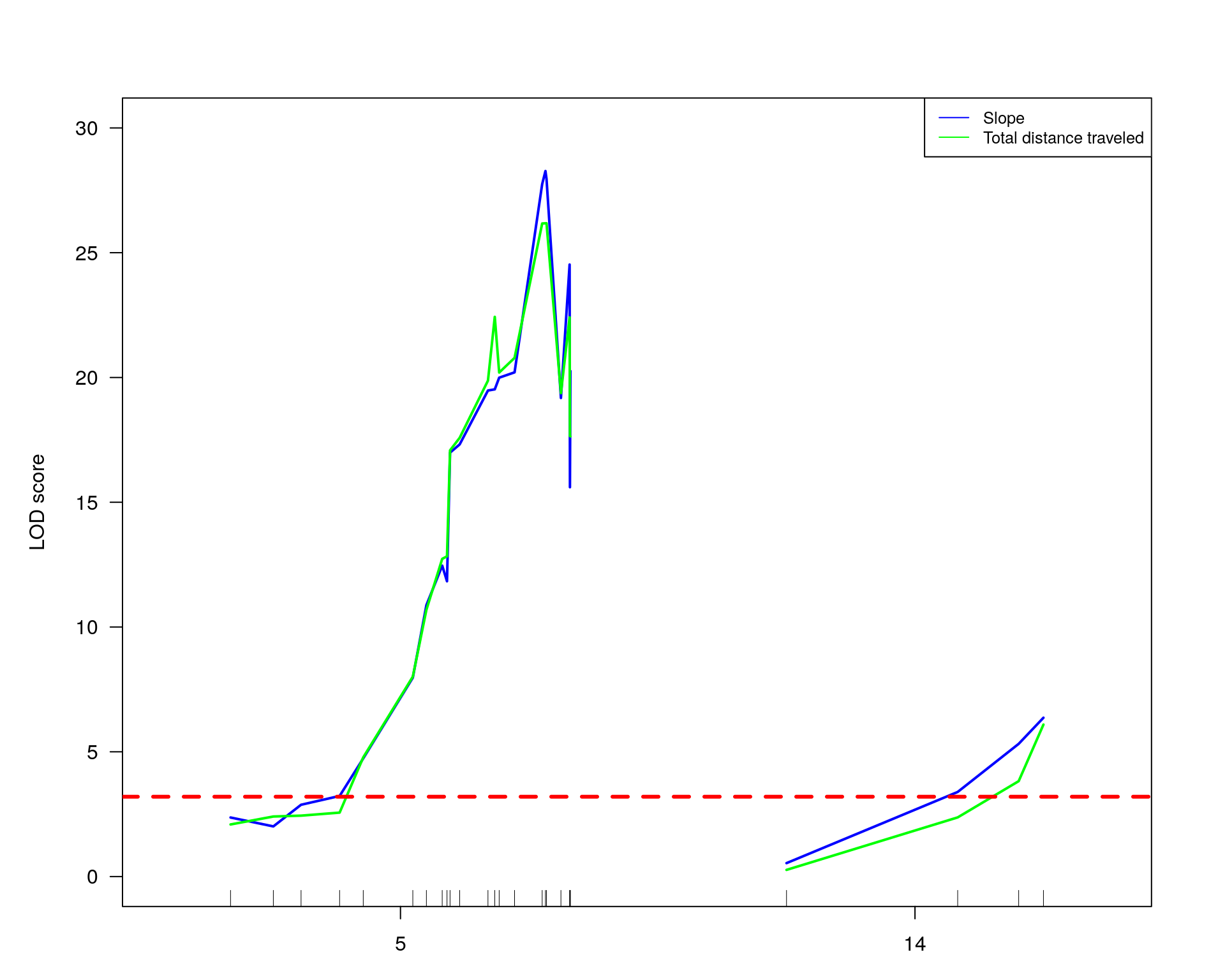
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
#Interval estimates of the location of QTL are commonly obtained via 1.5-LOD support intervals,
lodint.chr5.slope = lodint(out.mr[[2]], chr= 5)
lodint.chr5.slope chr pos lod
chr5-133052642-.-T-G 5 72.40547 20.20584
chr5-136332619-.-C-A 5 75.97770 28.27231
chr5-138956439-.-T-C 5 77.76853 19.17899lodint.chr14.slope = lodint(out.mr[[2]], chr= 14)
lodint.chr14.slope chr pos lod
chr14-50642778-.-G-A 14 26.14972 3.390345
chr14-69691477-.-G-T 14 36.07219 6.365103
chr14-69691477-.-G-T 14 36.07219 6.365103lodint.chr5.TotalDist3 = lodint(out.mr[[1]], chr= 5)
lodint.chr5.TotalDist3 chr pos lod
chr5-133052642-.-T-G 5 72.40547 20.78647
chr5-136332619-.-C-A 5 75.97770 26.18011
chr5-136844385-.-T-G 5 76.10078 26.18011
chr5-138956439-.-T-C 5 77.76853 19.37148lodint.chr14.TotalDist3 = lodint(out.mr[[1]], chr= 14)
lodint.chr14.TotalDist3 chr pos lod
chr14-62407765-.-T-A 14 33.20623 3.823005
chr14-69691477-.-G-T 14 36.07219 6.090514
chr14-69691477-.-G-T 14 36.07219 6.090514#loop for each pheno
for (i in 1:length(idx)){
print(colnames(WT144$pheno)[idx[[i]]])
#save plots
#operm_hist
pdf(paste0("output/", name,"-", colnames(WT144$pheno)[idx[[i]]], ".pdf"), width = 10, height = 10)
plot(operm[[i]][!is.infinite(operm[[i]])], main=paste(name, colnames(WT144$pheno)[idx[[i]]], sep="-"))
#mrscan_pheno
plot(out.mr[[i]], ylab=colnames(WT144$pheno)[idx[[i]]])
add.threshold(out.mr[[i]], perms = operm[[i]], alpha = 0.05, col="magenta")
#pxg
peak = summary(out.mr[[i]], threshold=summary(operm[[i]], alpha = 0.05)[[1]])
print(peak)
marker = c("chr5-136332619-.-C-A", "chr14-69691477-.-G-T")
for (mar in marker){
par(mar=c(3, 5, 4, 3))
plotPXG(WT144, marker=mar, jitter = 0.25, pheno.col = idx[[i]], infer=F, main=paste(mar),
mgp=c(3.4,1,0))
}
#interaction_effect
effectplot(WT144,
mname1 = rownames(peak)[1],
mname2 = rownames(peak)[2],
pheno.col = idx[[i]], add.legend=T, legend.lab = "")
##Multiple-QTL analyses
# After performing the single- and two-QTL genome scans, it’s best to bring the identified loci together into a joint model, which we then refine and from which we may explore the possibility of further QTL. In this effort, we work with “QTL objects” created by makeqtl(). We fit multiple-QTL models with fitqtl(). A number of additional functions will be introduced below.
#First, we create a QTL object containing the loci on chr 5 and 14.
#chr5-136332619-.-C-A 75.97770
#chr14-69691477-.-G-T 36.07219
WT144 <- sim.geno(WT144, n.draws=64)
qtl <- makeqtl(WT144, chr=c(5,14), pos=c(75.97770, 36.07219), what="draws")
out.fq <- fitqtl(WT144, pheno.col=idx[[i]], qtl=qtl, method = "hk")
print(summary(out.fq))
#We may obtain the estimated effects of the QTL via get.ests=TRUE. We use dropone=FALSE to suppress the drop-one-term analysis.
print(summary(fitqtl(WT144, pheno.col=idx[[i]], qtl=qtl, method="hk", get.ests=TRUE, dropone=FALSE)))
#To assess the possibility of an interaction between the two QTL, we may fit the model with the interaction, indicated via a model “formula”.
print("To assess the possibility of an interaction between the two QTL")
out.fqi <- fitqtl(WT144, pheno.col=idx[[i]], qtl=qtl, method="hk", formula=y~Q1+Q2+Q1:Q2)
print(summary(out.fqi))
#complete_scan2
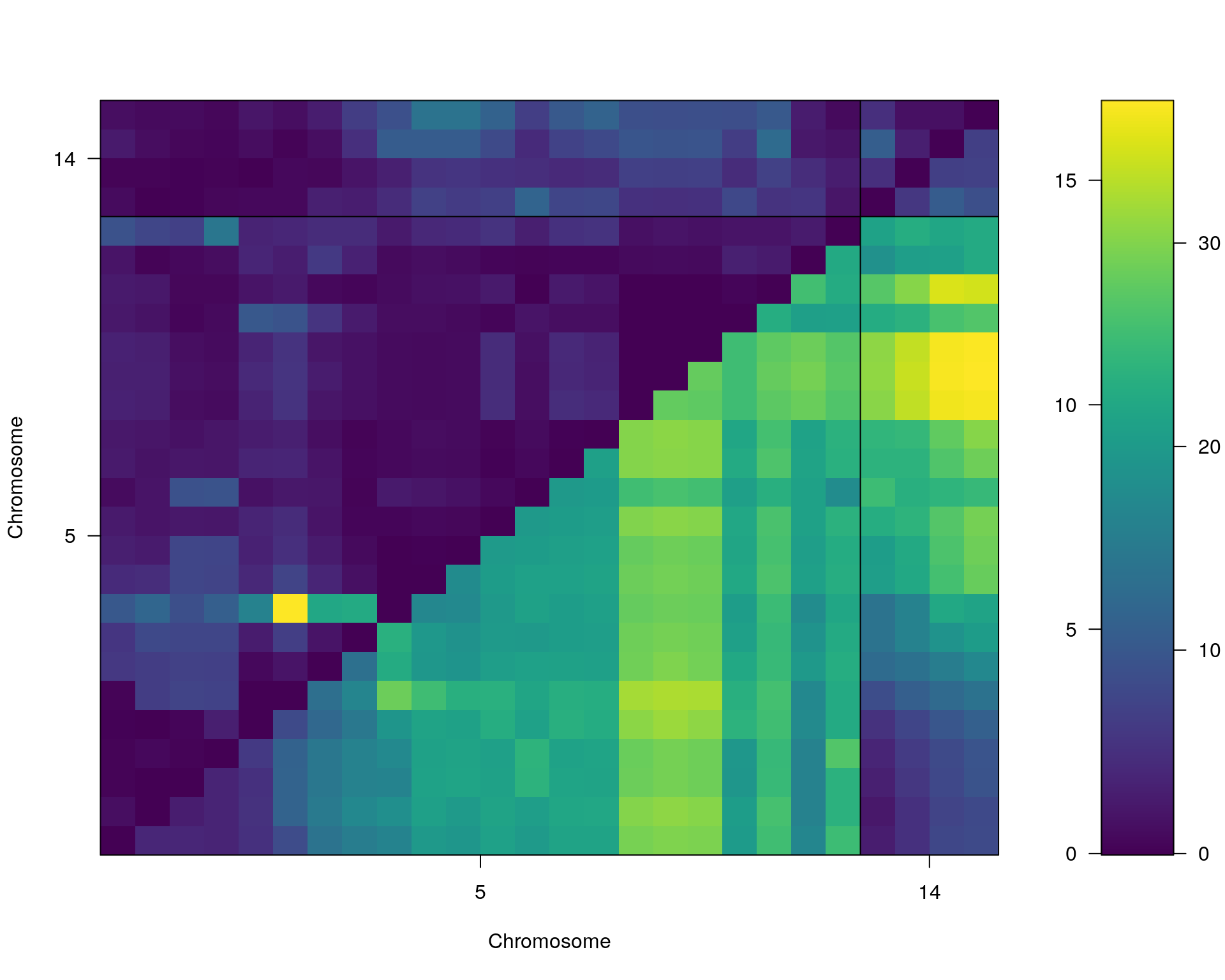
plot(st[[i]])
#toptwo_scan2
plot(st[[i]], chr = summary(out.mr[[i]])[order(-summary(out.mr[[i]])$lod), "chr"][1:2])
dev.off()
}[1] "TotalDist_3" chr pos lod
chr5-136844385-.-T-G 5 76.1 26.18
chr14-69691477-.-G-T 14 36.1 6.09
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 281
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 3170972617 792743154 31.45631 40.28112 0 0
Error 276 4701133507 17033092
Total 280 7872106124
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 2 2.423e+09 25.367 30.783 71.13 0 < 2e-16 ***
14@36.1 2 4.478e+08 5.552 5.689 13.15 0 3.52e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 281
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 3170972617 792743154 31.45631 40.28112 0 0
Error 276 4701133507 17033092
Total 280 7872106124
Estimated effects:
-----------------
est SE t
Intercept 7106.2 251.6 28.249
5@76.0a 3548.9 349.2 10.162
5@76.0d -2908.4 497.5 -5.846
14@36.1a 1818.8 380.1 4.785
14@36.1d -1335.6 503.4 -2.653
[1] "To assess the possibility of an interaction between the two QTL"
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 281
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q1:Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 8 3558447363 444805920 36.70494 45.20324 0 0
Error 272 4313658761 15859040
Total 280 7872106124
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 6 2.811e+09 30.615 35.705 29.539 0 < 2e-16 ***
14@36.1 6 8.353e+08 10.801 10.611 8.778 0 9.41e-09 ***
5@76.0:14@36.1 4 3.875e+08 5.249 4.922 6.108 0 0.000101 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1[1] "Slope" chr pos lod
chr5-136332619-.-C-A 5 76.0 28.27
chr14-69691477-.-G-T 14 36.1 6.37
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 309
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 464846.8 116211.711 32.78381 38.65106 0 0
Error 304 737828.6 2427.068
Total 308 1202675.5
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 2 355665 26.398 29.573 73.27 0 < 2e-16 ***
14@36.1 2 55228 4.843 4.592 11.38 0 1.72e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 309
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 4 464846.8 116211.711 32.78381 38.65106 0 0
Error 304 737828.6 2427.068
Total 308 1202675.5
Estimated effects:
-----------------
est SE t
Intercept -26.045 2.858 -9.113
5@76.0a 40.839 3.926 10.402
5@76.0d -33.296 5.694 -5.848
14@36.1a 19.766 4.308 4.588
14@36.1d -11.540 5.726 -2.015
[1] "To assess the possibility of an interaction between the two QTL"
fitqtl summary
Method: multiple imputation
Model: normal phenotype
Number of observations : 309
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q1:Q2
df SS MS LOD %var Pvalue(Chi2) Pvalue(F)
Model 8 507426.4 63428.302 36.77224 42.19147 0 0
Error 300 695249.1 2317.497
Total 308 1202675.5
Drop one QTL at a time ANOVA table:
----------------------------------
df Type III SS LOD %var F value Pvalue(Chi2) Pvalue(F)
5@76.0 6 398245 30.386 33.113 28.640 0.000 < 2e-16 ***
14@36.1 6 97808 8.832 8.133 7.034 0.000 5.11e-07 ***
5@76.0:14@36.1 4 42580 3.988 3.540 4.593 0.001 0.0013 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Plot the Dist_vs_age separate by genotype for each marker
plot_graph <- function(cross, name){
# Plot the Dist vs age, separate by genotype for each marker
allphen <- cross$pheno
allgeno <- as.data.frame(pull.geno(cross))
rownames(allgeno) <- cross$pheno$AnimalName
allphen <- merge(allphen, allgeno, by.x="AnimalName", by.y="row.names", all.x=TRUE)
pdf(paste0("output/", name, "_TotalDist_vs_TestAge_by_marker.pdf"))
for (mar in colnames(allgeno)){
subplot <- allphen[!is.na(allphen[,mar, drop=FALSE]),] %>%
pivot_longer(cols = starts_with("TestAge_"), names_to="test.no", values_to = "Age") %>%
separate(test.no, c("empty1", "test1")) %>%
pivot_longer(cols = starts_with("TotalDist_"), names_to="test.no.2", values_to = "Dist") %>%
separate(test.no.2, c("empty2", "test2")) %>% filter(test1==test2)
subplot <- as.data.frame(subplot)
subplot[, mar] <- factor(subplot[, mar, drop=T])
levels(subplot[,mar]) <- c("AA","AB","BB")
p <- ggplot(subplot, aes(Age, Dist, color=get(mar), group=get(mar))) +
geom_point() +
labs(color = "Genotype") +
scale_color_brewer(palette = "Set1") +
stat_smooth(method="lm", show.legend = FALSE, formula = y~x) +
labs(title=paste0("effect for marker ", mar))
print(p)
}
dev.off()
}
plot_int_graph <- function(cross, name, basem){
# Plot the Dist vs age, separate by genotype for each marker
allphen <- cross$pheno
allgeno <- as.data.frame(pull.geno(cross))
rownames(allgeno) <- cross$pheno$AnimalName
allphen <- merge(allphen, allgeno, by.x="AnimalName", by.y="row.names", all.x=TRUE)
#pdf(paste0("output/", name, ".pdf"))
for (mar in colnames(allgeno)){
print(mar)
subplot <- allphen[!is.na(allphen[,mar, drop=FALSE]),] %>%
pivot_longer(cols = starts_with("TestAge_"), names_to="test.no", values_to = "Age") %>%
separate(test.no, c("empty1", "test1")) %>%
pivot_longer(cols = starts_with("TotalDist_"), names_to="test.no.2", values_to = "Dist") %>%
separate(test.no.2, c("empty2", "test2")) %>% filter(test1==test2)
subplot <- as.data.frame(subplot)
subplot$interactions <- factor(subplot[, basem] + subplot[, mar]*3)
# 1:1 -> 4 1:2 -> 7 1:3 -> 10
# 2:1 -> 5 2:2 -> 8 2:3 -> 11
# 3:1 -> 6 3:2 -> 9 3:3 -> 12
itypes <- c(1,2,3,"AA:AA", "AB:AA", "BB:AA",
"AA:AB", "AB:AB", "BB:AB",
"AA:BB", "AB:BB", "BB:BB")
levels(subplot$interactions) <- itypes[as.integer(levels(subplot$interactions))]
#replaces 1 2 and 3 as AA, AB and BB
subplot[, basem][subplot[, basem] == 1] <- "AA"
subplot[, basem][subplot[, basem] == 2] <- "AB"
subplot[, basem][subplot[, basem] == 3] <- "BB"
subplot[, mar][subplot[, mar] == 1] <- "AA"
subplot[, mar][subplot[, mar] == 2] <- "AB"
subplot[, mar][subplot[, mar] == 3] <- "BB"
p1 <- ggplot(subplot, aes(Age, Dist, color=interactions, group=interactions)) +
geom_point(shape = 20) +
#scale_color_discrete("Genotype") +
labs(color = "Genotype") +
ylab("TotalDist") +
xlab("TestAge") +
#scale_color_brewer(palette = "Set1") +
scale_color_manual(breaks = c("AA:AA", "AB:AA", "BB:AA", "AA:AB",
"AB:AB", "BB:AB", "AA:BB", "AB:BB", "BB:BB"),
#values = RColorBrewer::brewer.pal(9, "Set1")[9:1]) +
values = c("#999999", "#F781BF", "#A65628","#9a9a00", "#FF7F00", "#984EA3", "#4DAF4A", "#377EB8", "#E41A1C")) +
stat_smooth(method="lm", show.legend = FALSE, formula = y~x) +
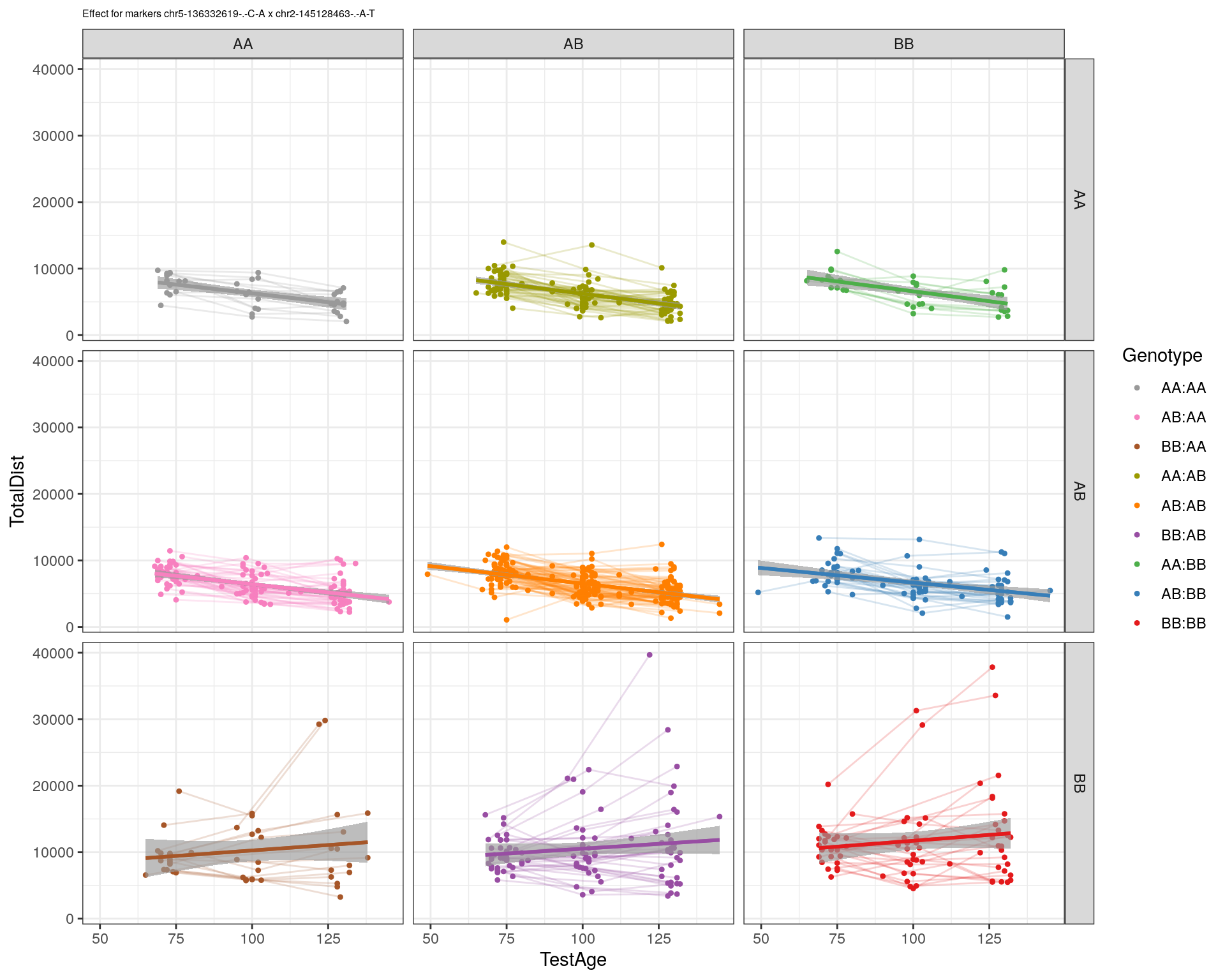
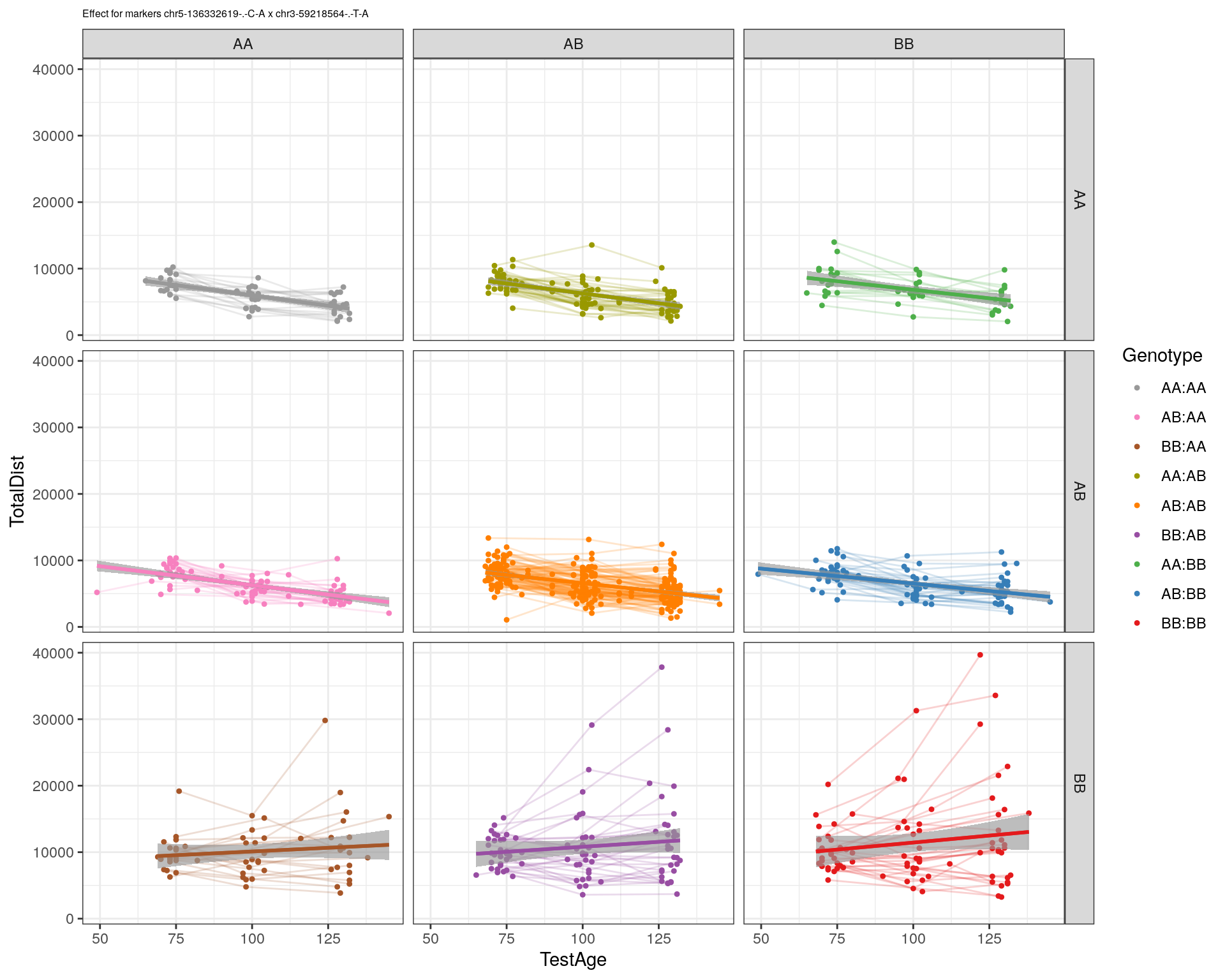
labs(title=paste0("effect for markers ", basem, " x ", mar)) +
theme_bw()
print(p1)
p2 <- ggplot(subplot, aes(Age, Dist, color=interactions, group=interactions)) +
geom_point(shape = 20) +
geom_line(aes(group = AnimalName, color = interactions), show.legend = FALSE, alpha = 0.2) +
geom_smooth(method='lm', formula= y~x, show.legend = FALSE) +
#scale_color_discrete("Genotype") +
labs(color = "Genotype") +
ylab("TotalDist") +
xlab("TestAge") +
#scale_color_brewer(palette = "Set1") +
scale_color_manual(breaks = c("AA:AA", "AB:AA", "BB:AA", "AA:AB",
"AB:AB", "BB:AB", "AA:BB", "AB:BB", "BB:BB"),
#values = RColorBrewer::brewer.pal(9, "Set1")[9:1]) +
values = c("#999999", "#F781BF", "#A65628","#9a9a00", "#FF7F00", "#984EA3", "#4DAF4A", "#377EB8", "#E41A1C")) +
stat_smooth(method="lm", show.legend = FALSE, formula = y~x) +
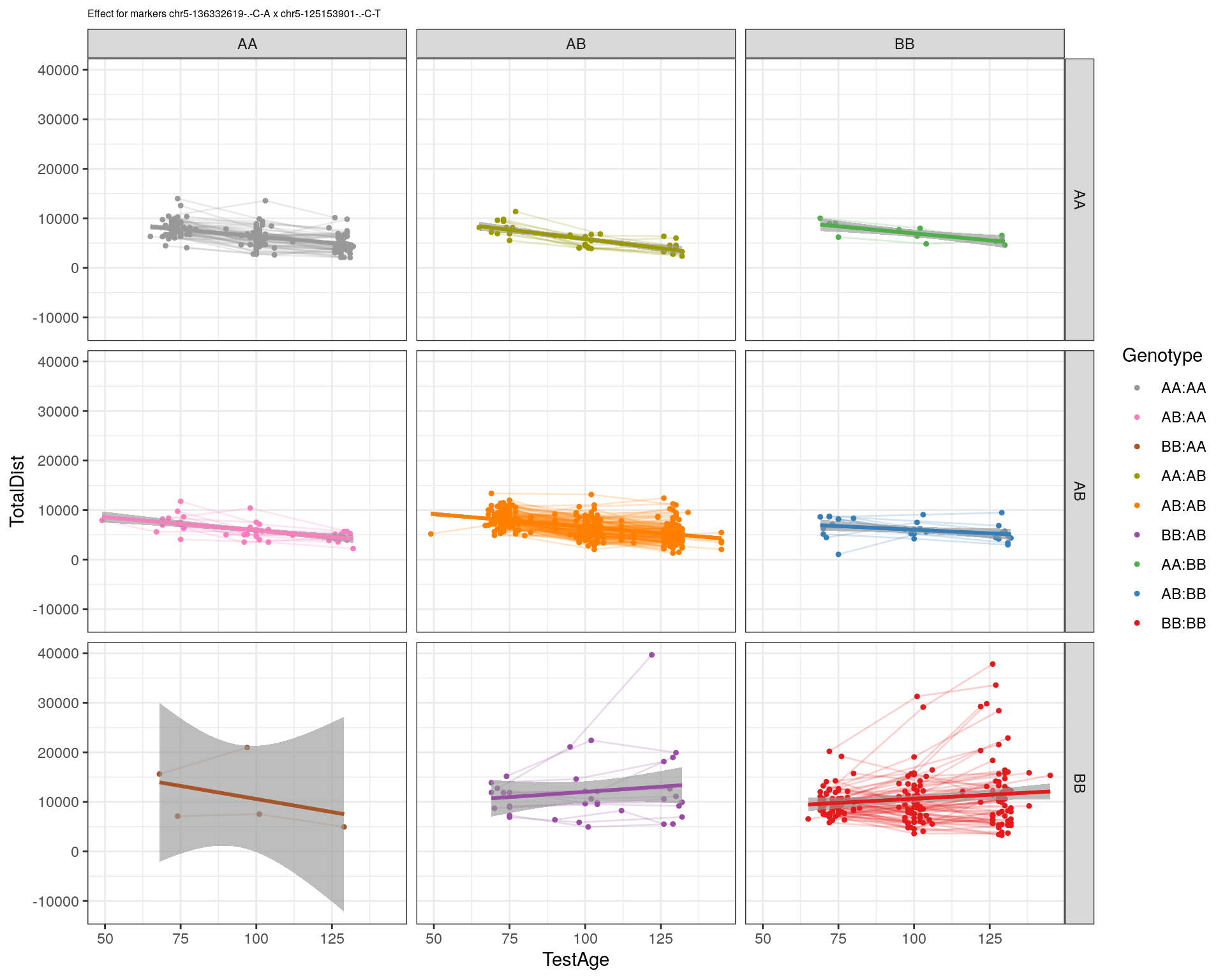
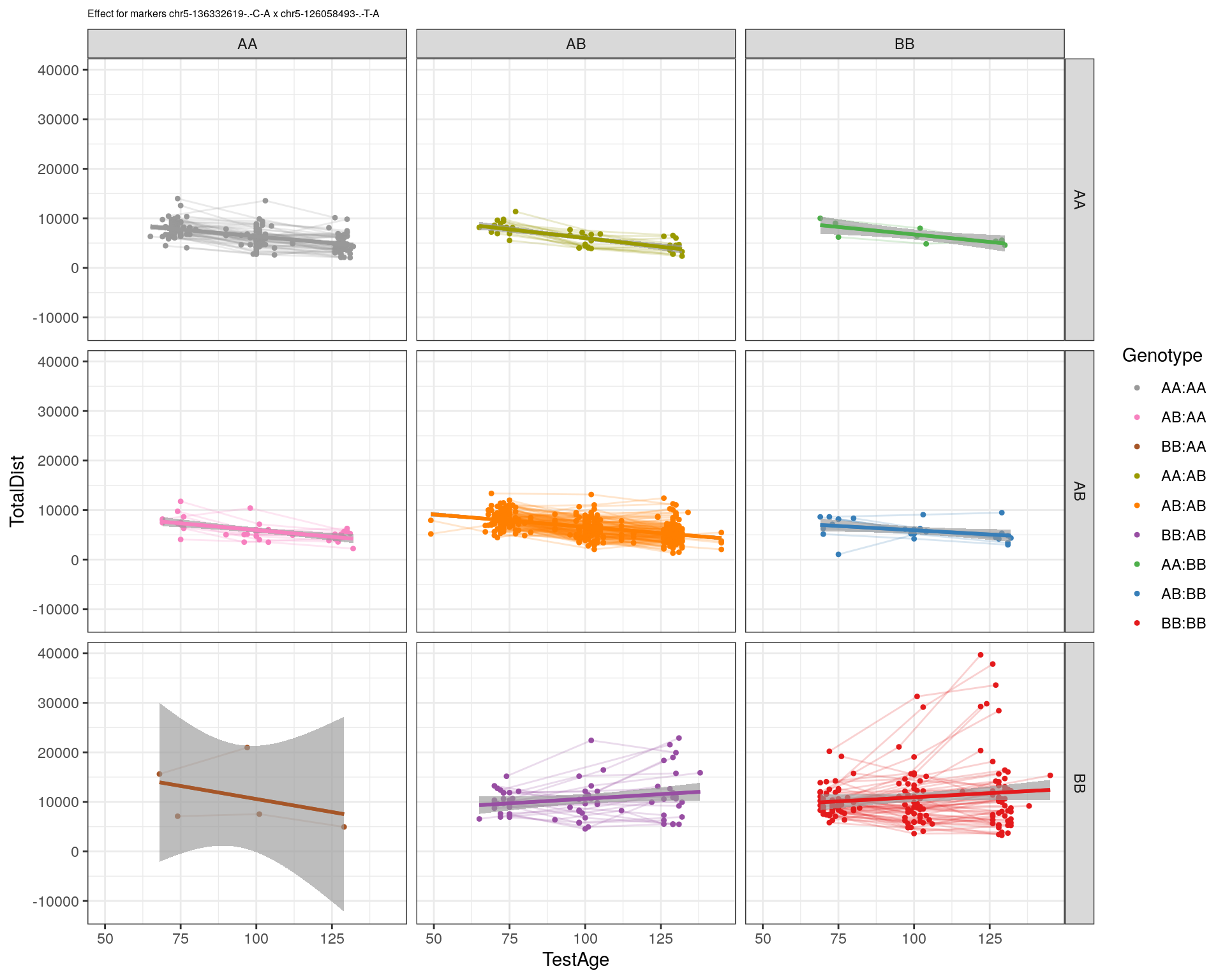
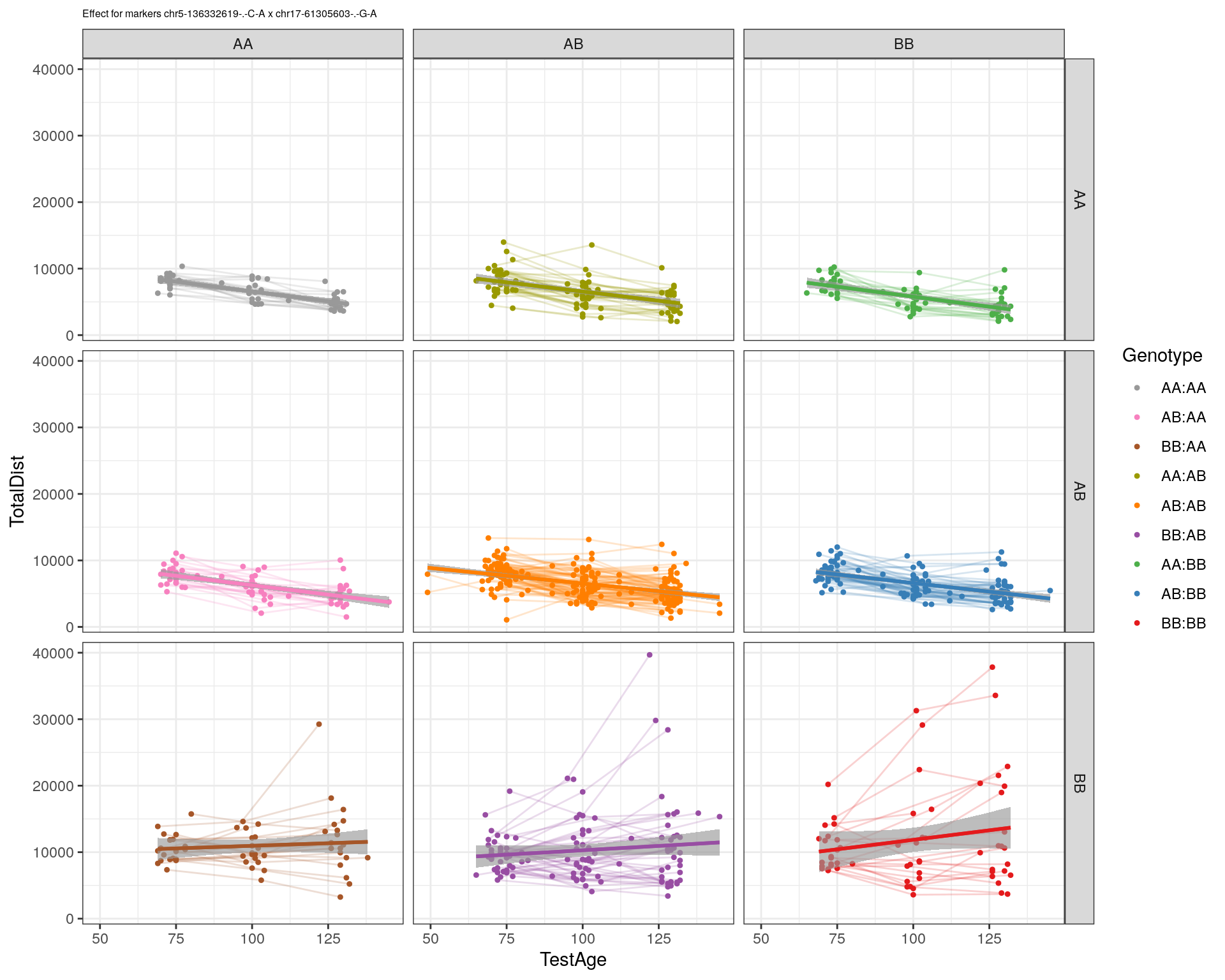
labs(title=paste0("Effect for markers ", basem, " x ", mar)) +
facet_grid(subplot[, basem] ~ subplot[, mar]) +
theme_bw() +
theme(plot.title = element_text(size=6))
print(p2)
}
#dev.off()
}
plot_graph(WT144, "WT144_effect_for_marker_plots")png
2 plot_int_graph(WT144, paste0("WT144_effect_for_marker_", "_int_chr5-136332619-.-C-A"), "chr5-136332619-.-C-A")[1] "chr1-65643621-.-C-A"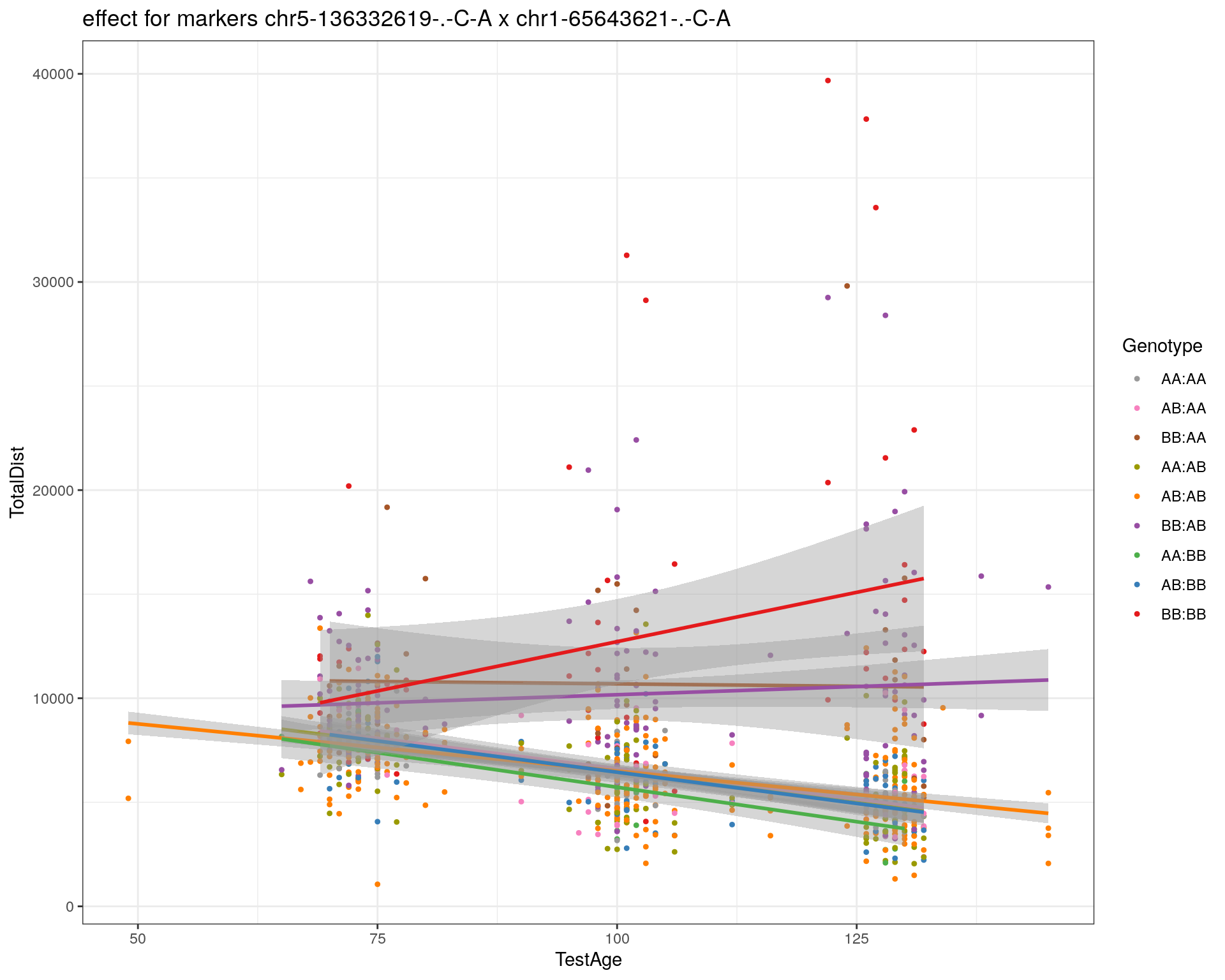
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
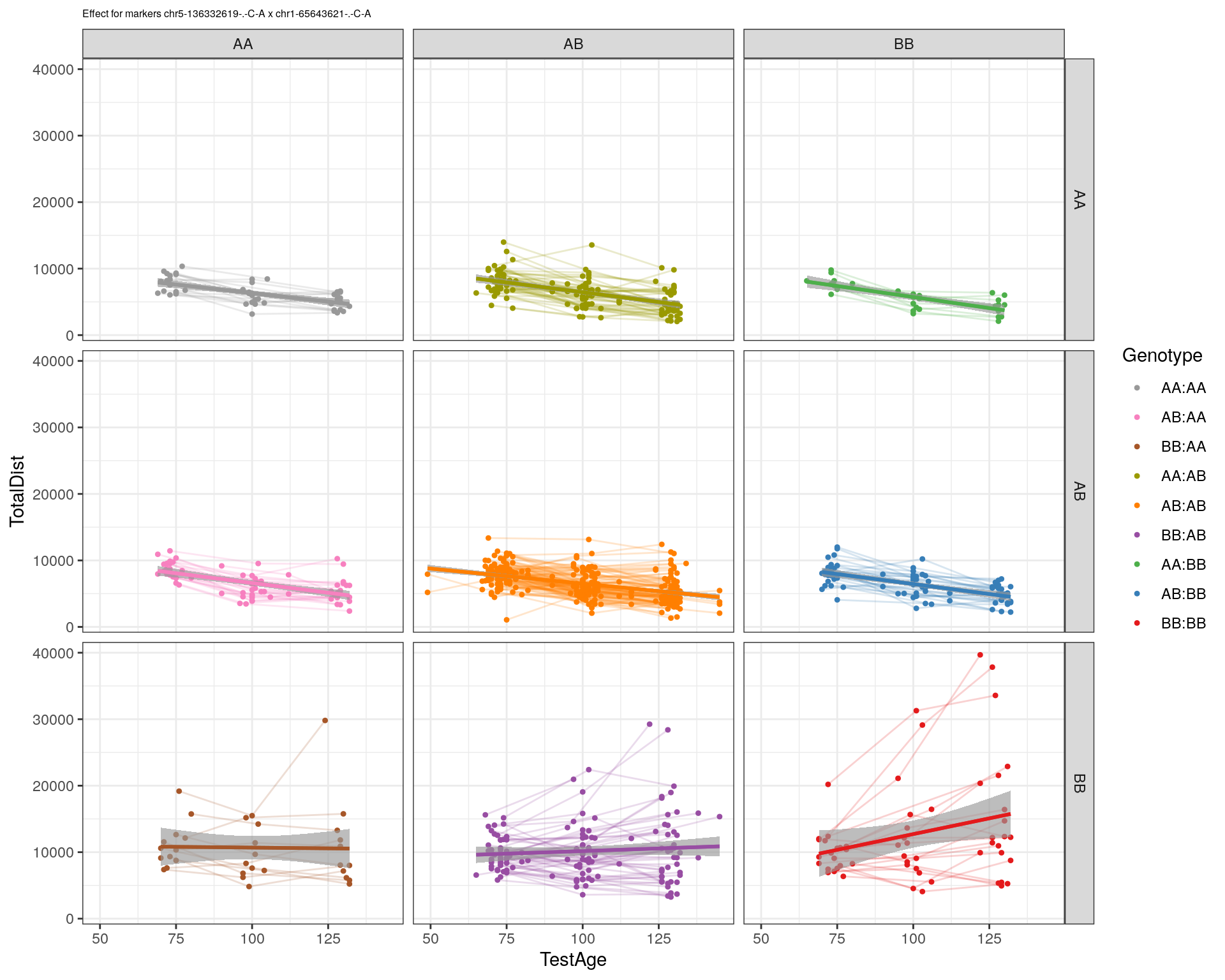
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr1-74679418-.-A-G"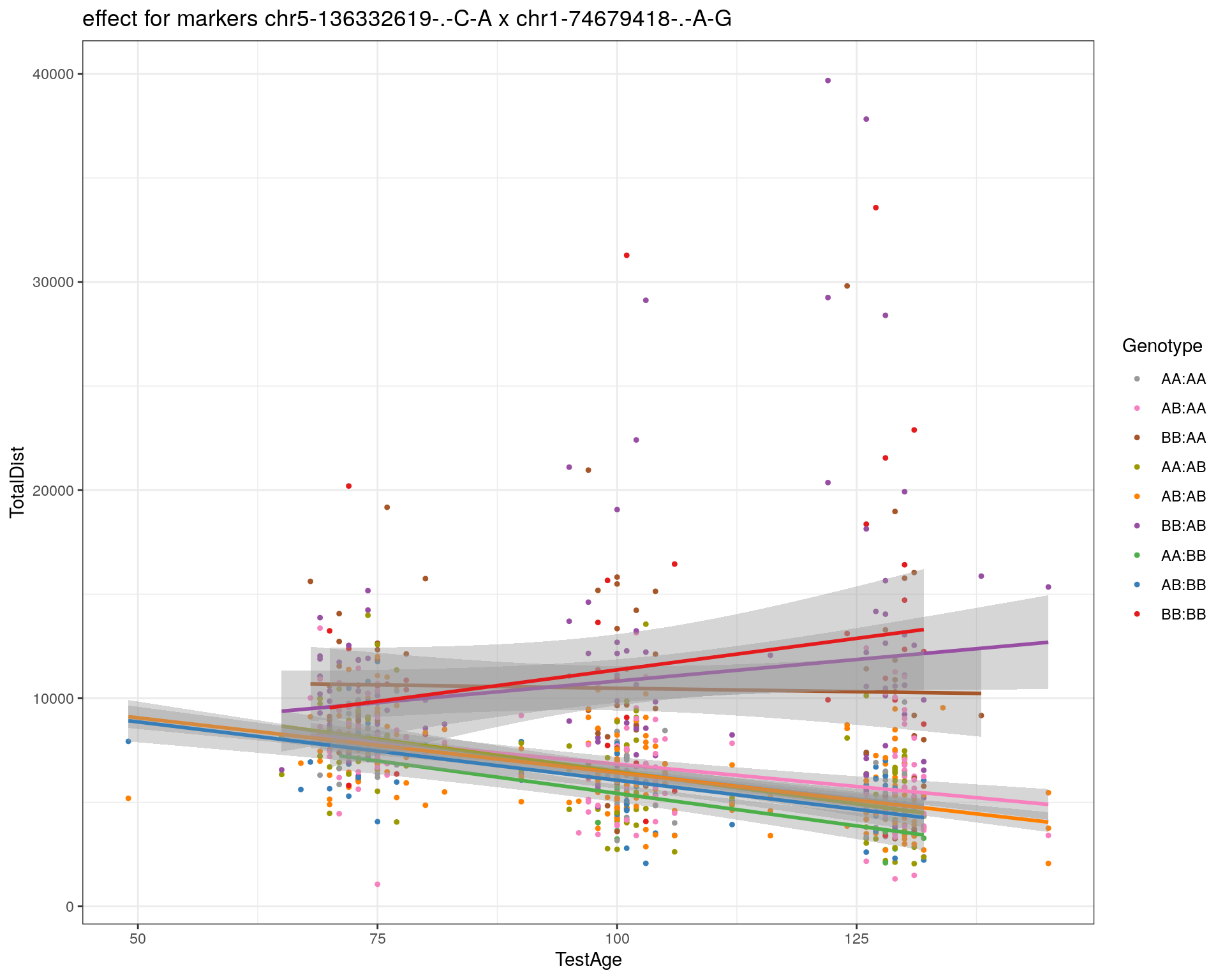
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
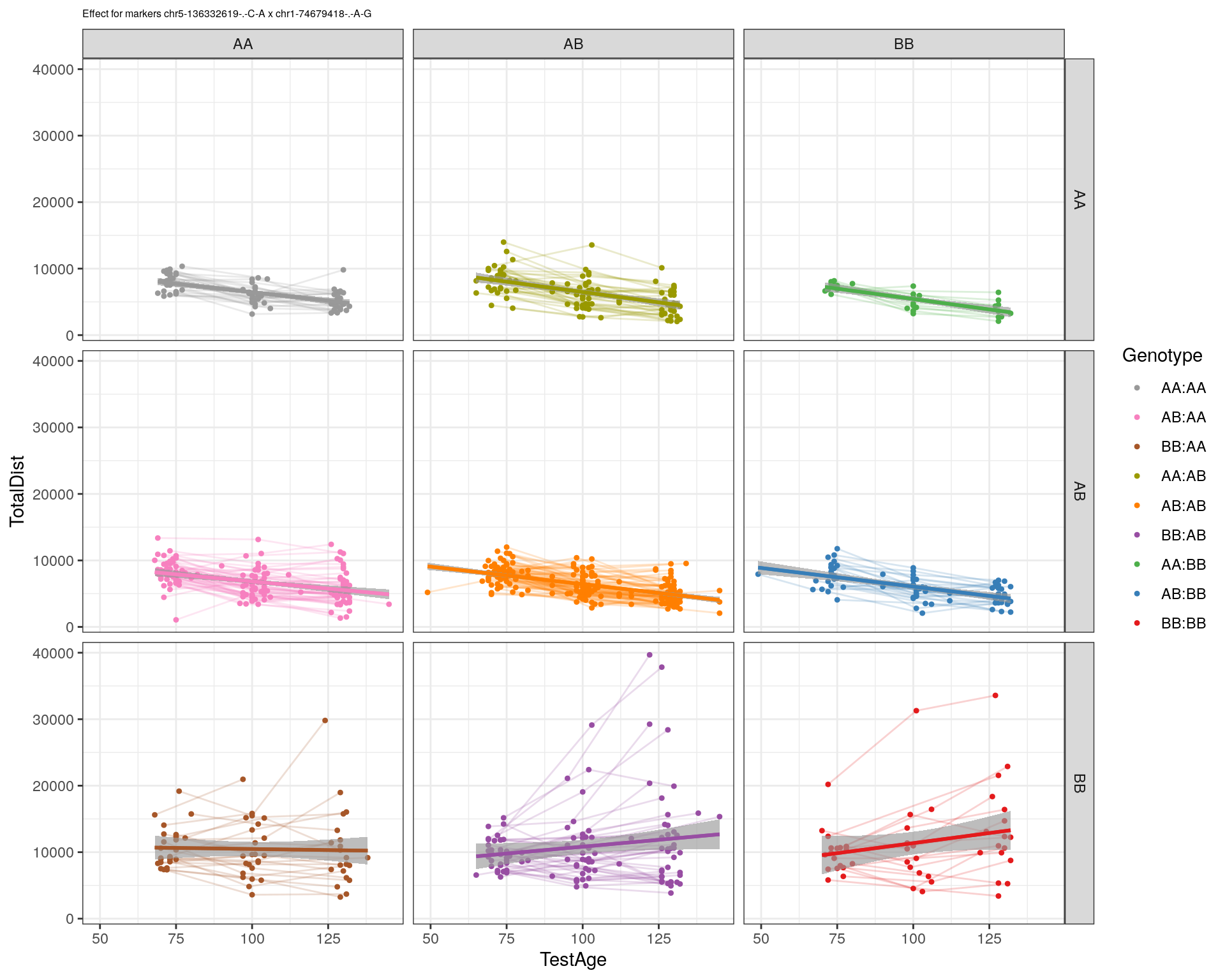
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr1-79823528-.-A-G"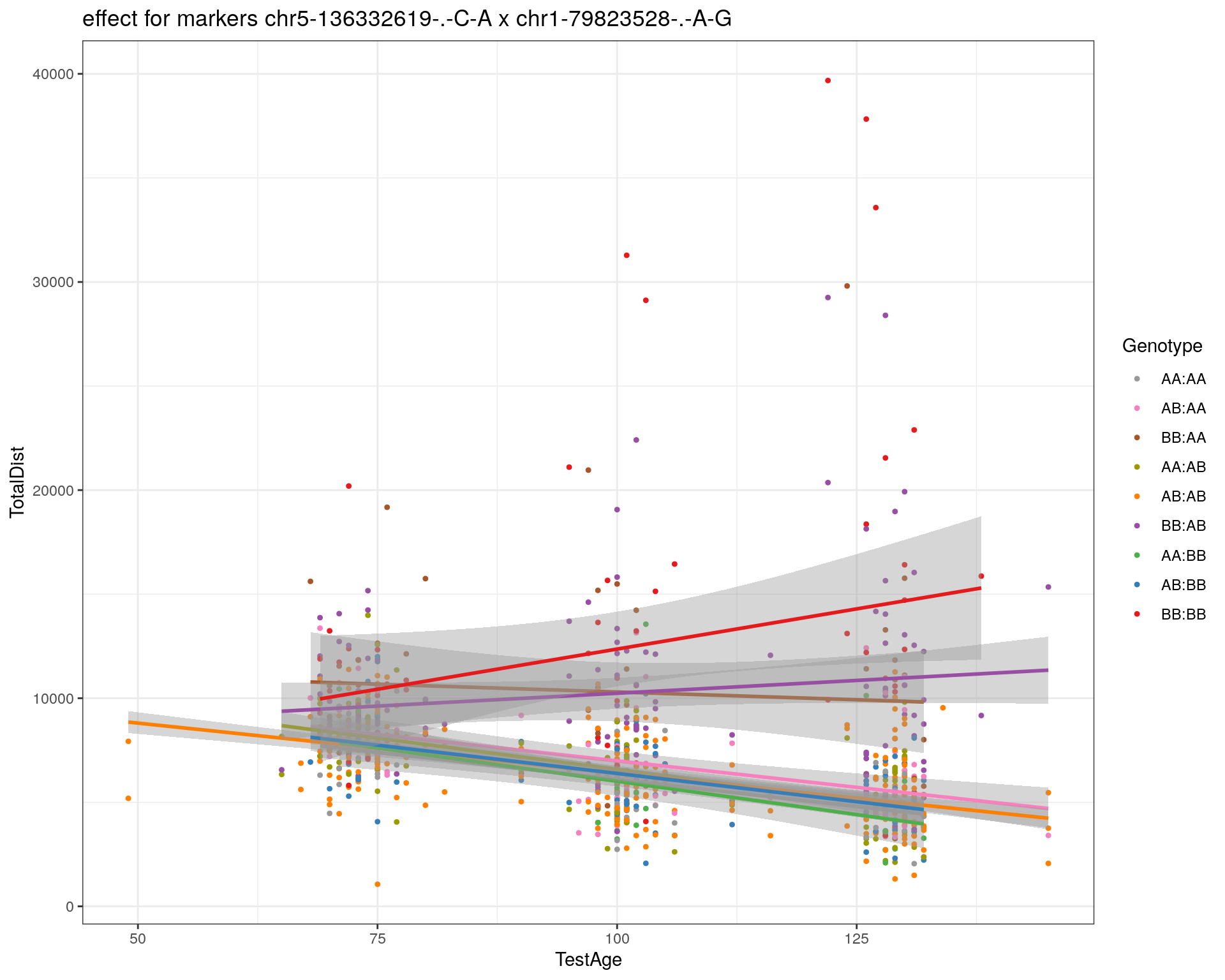
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
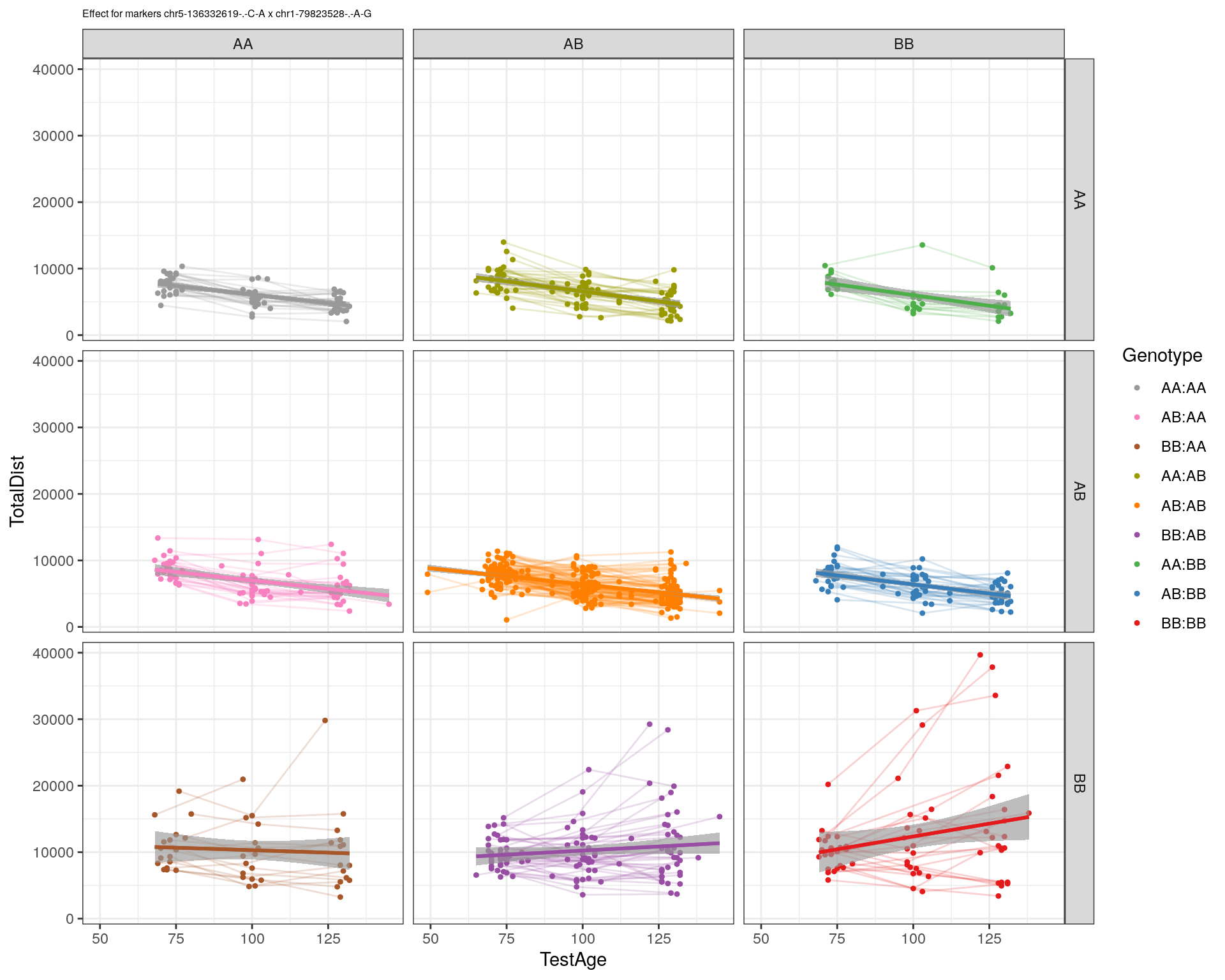
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr1-86851039-.-A-G"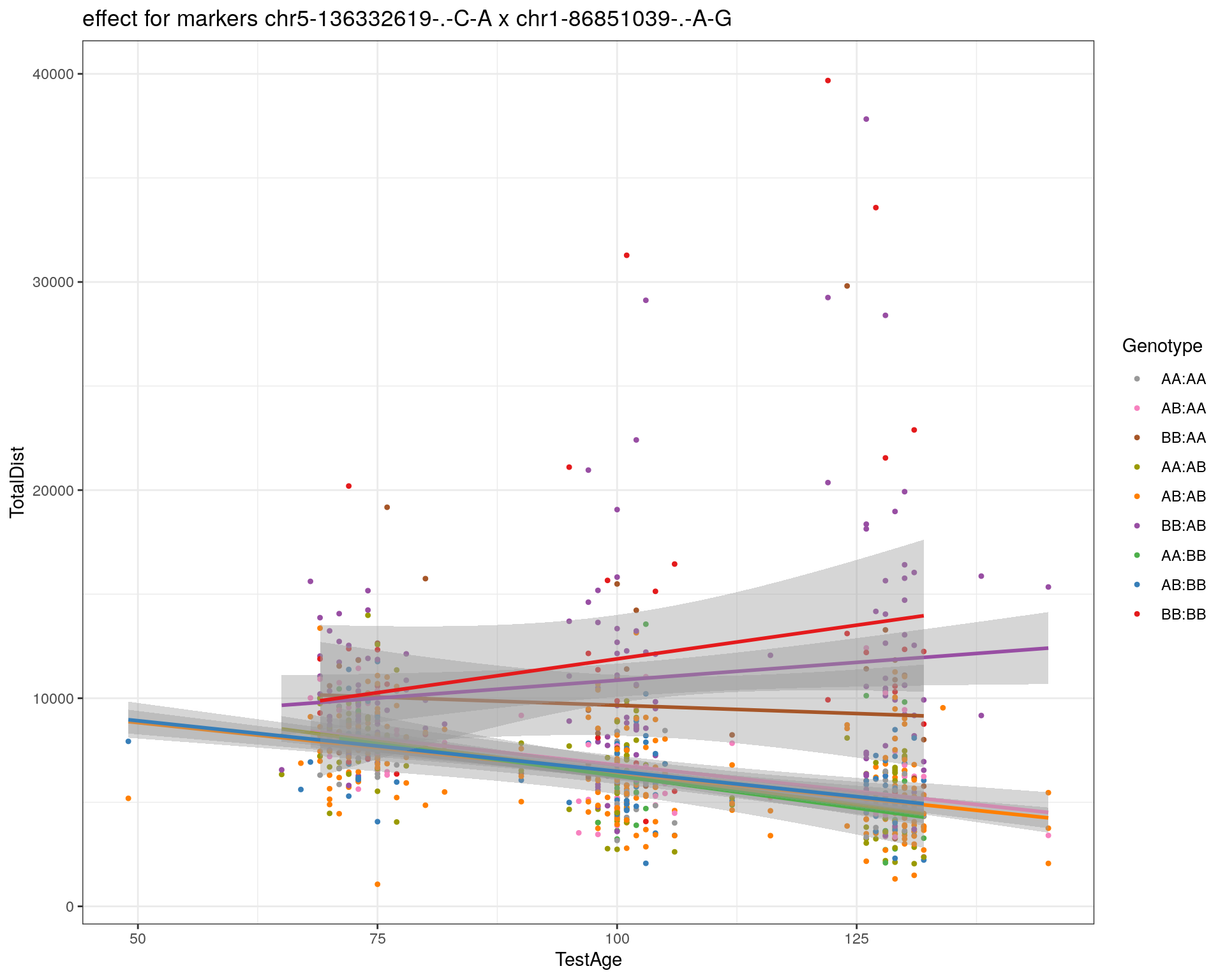
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
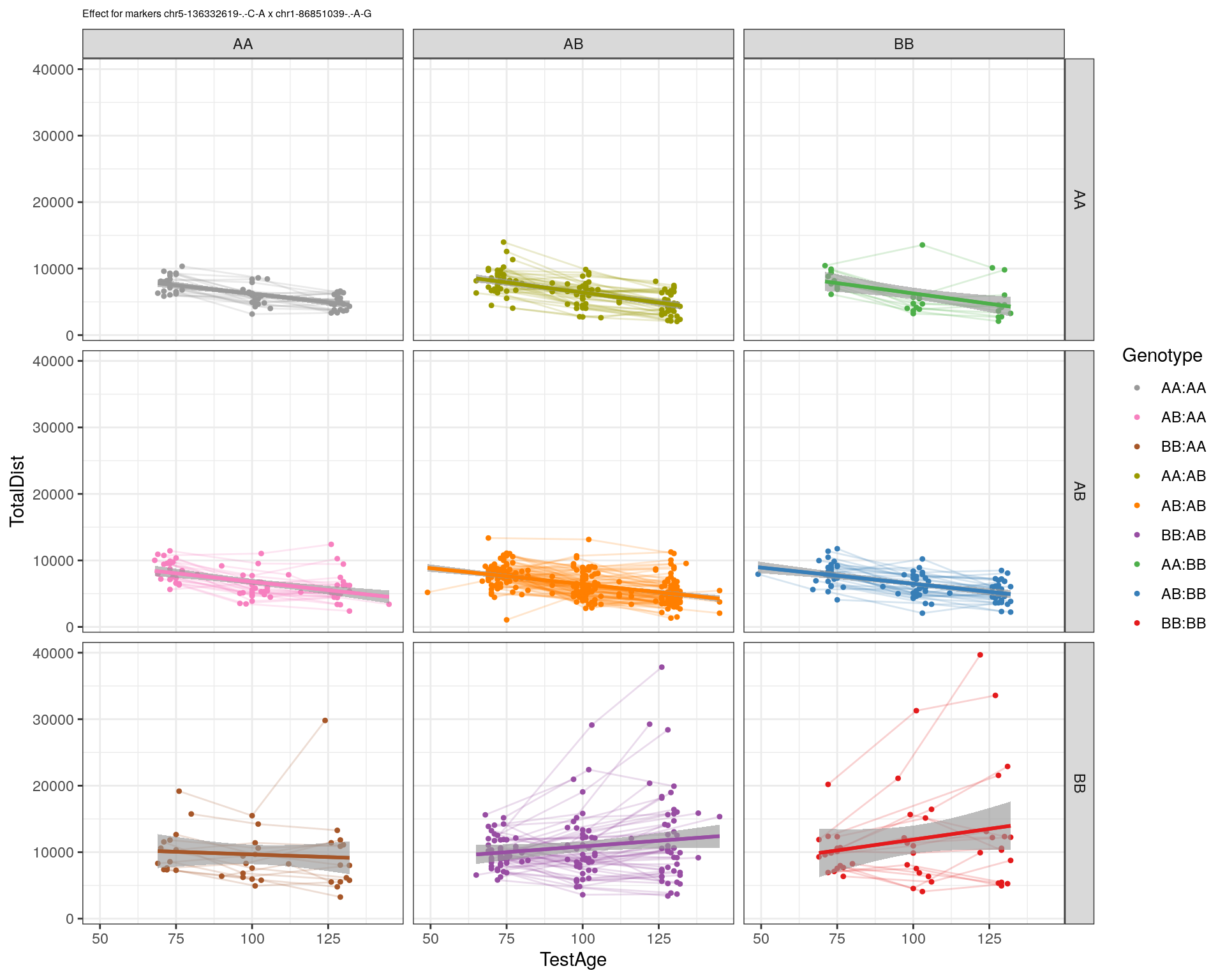
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr1-188155143-.-G-A"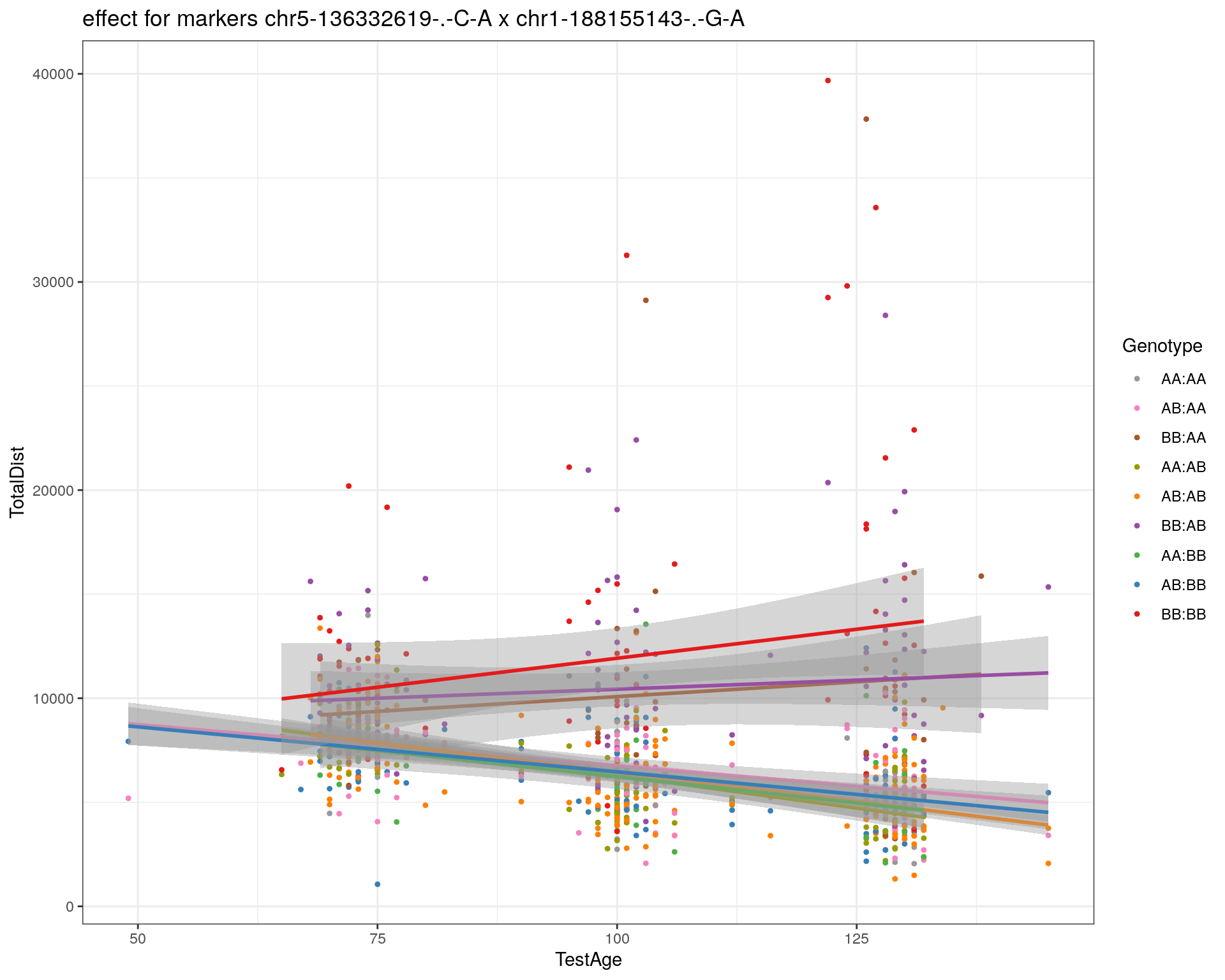
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
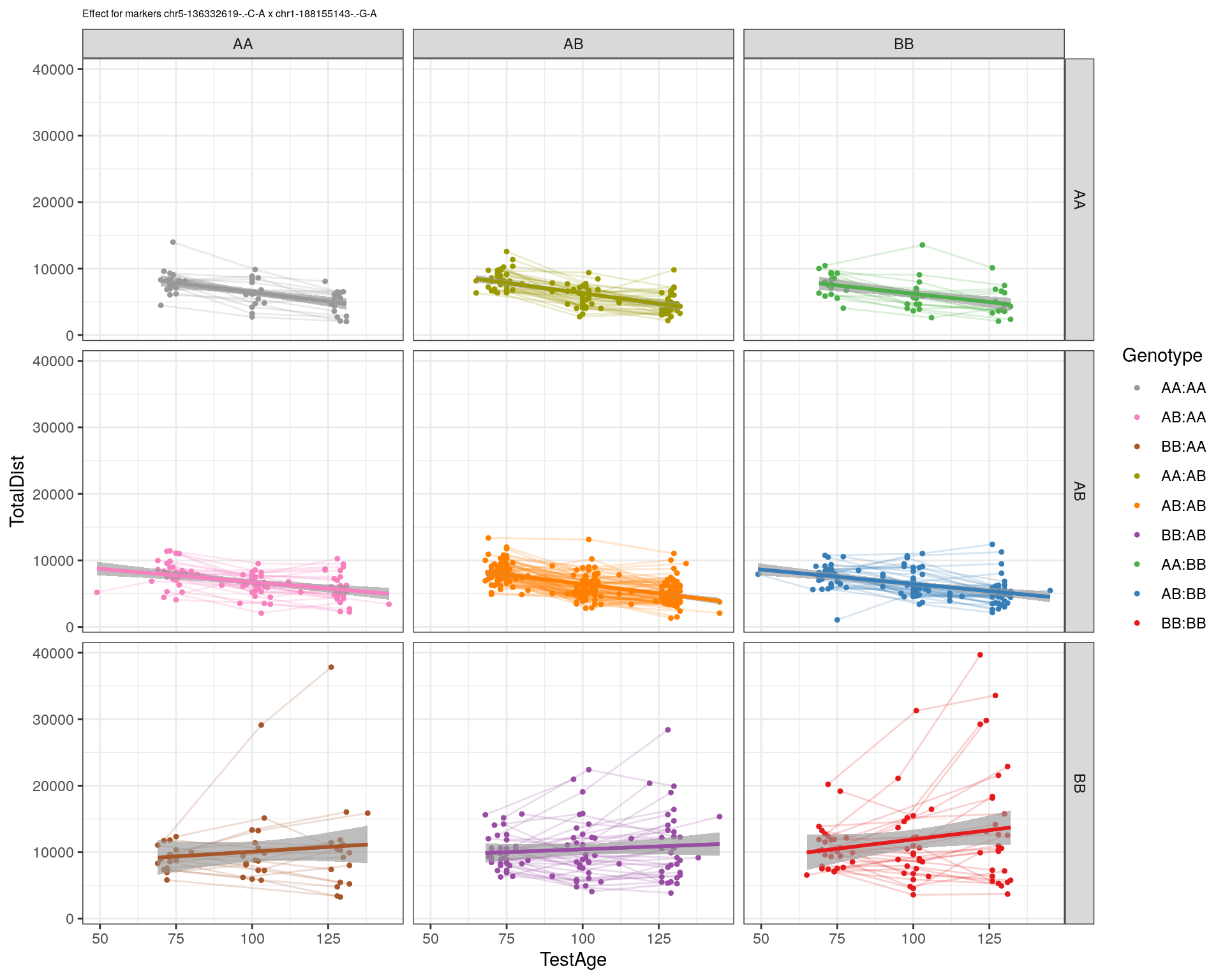
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr2-7018694-.-G-T"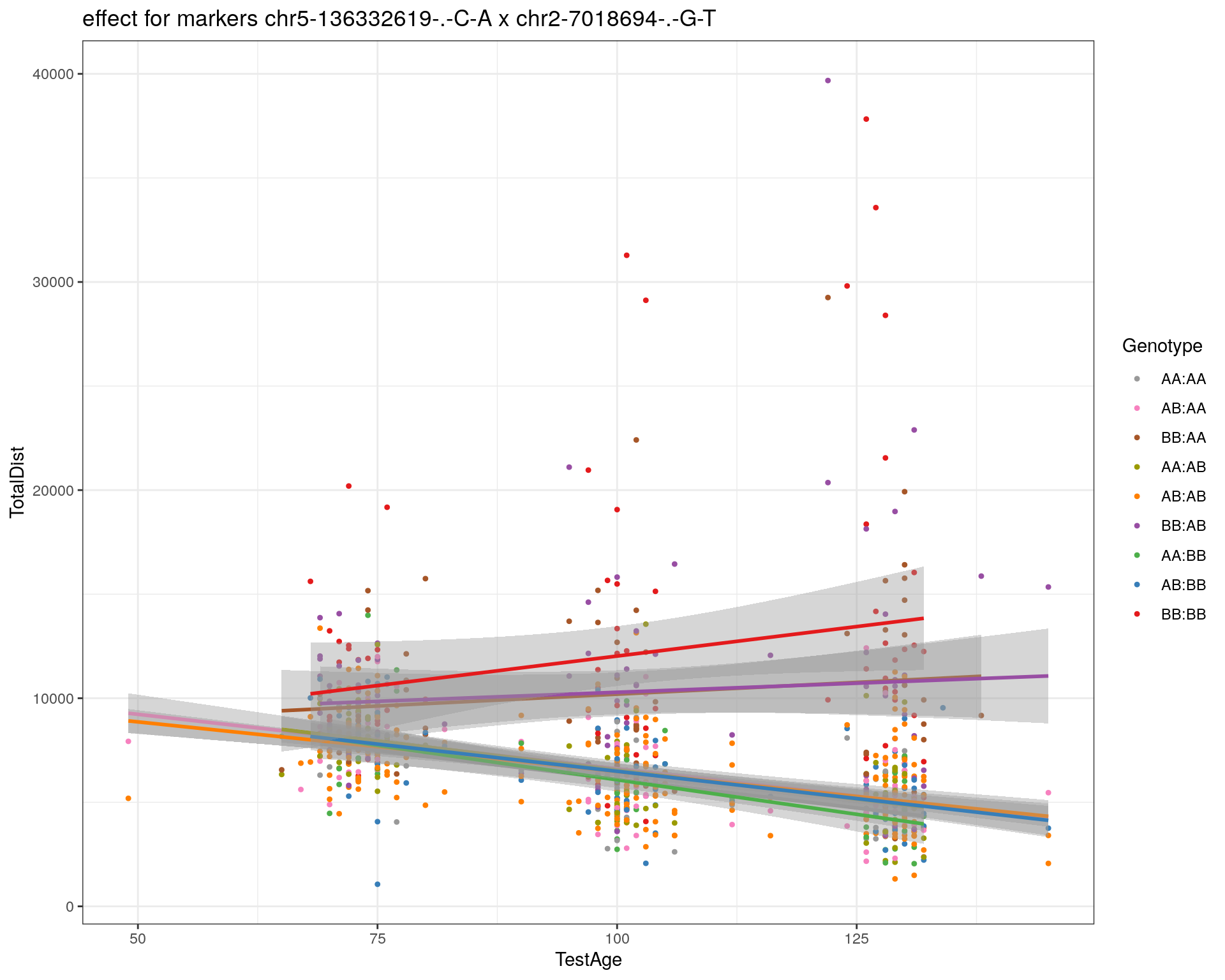
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
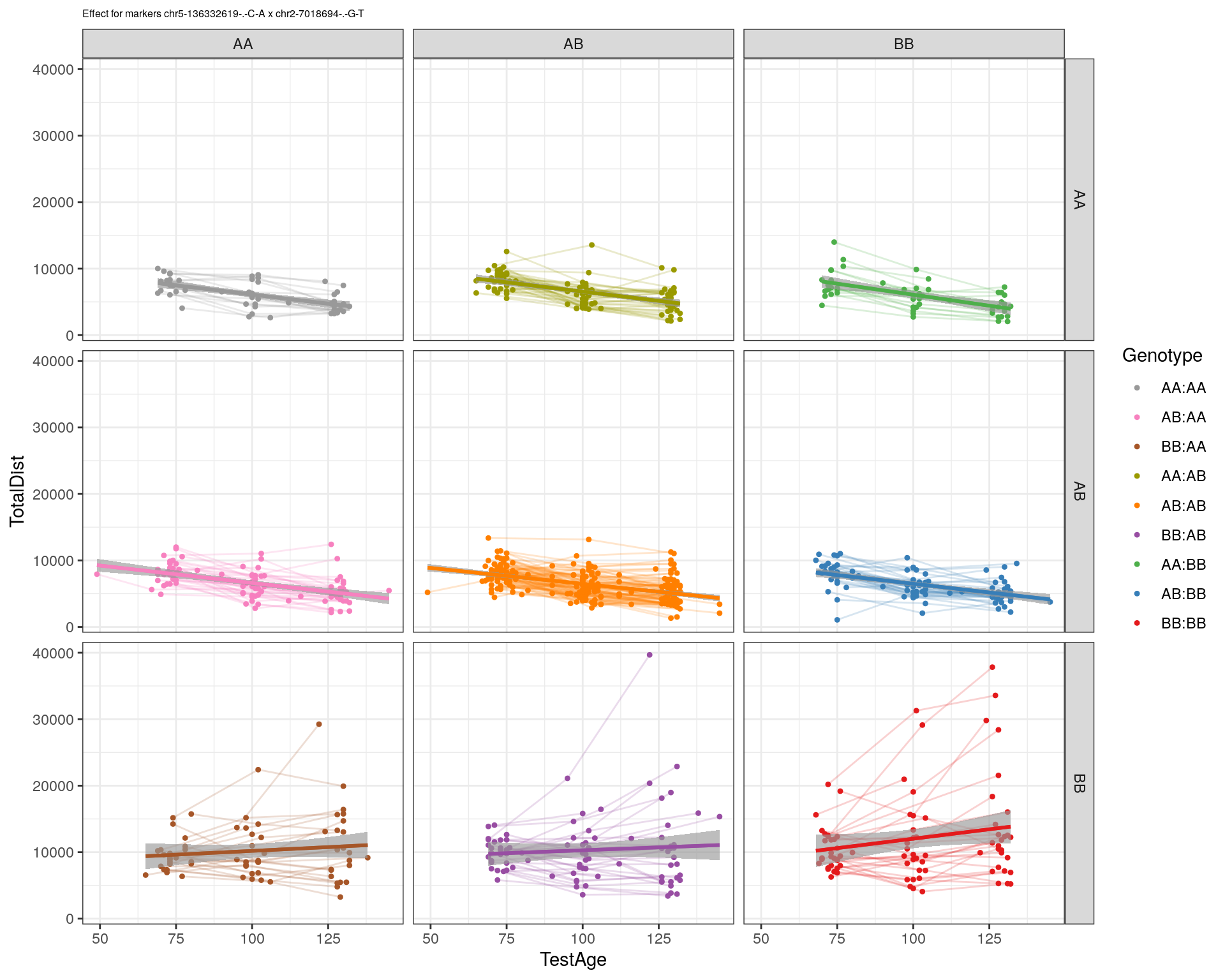
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr2-79706301-.-T-A"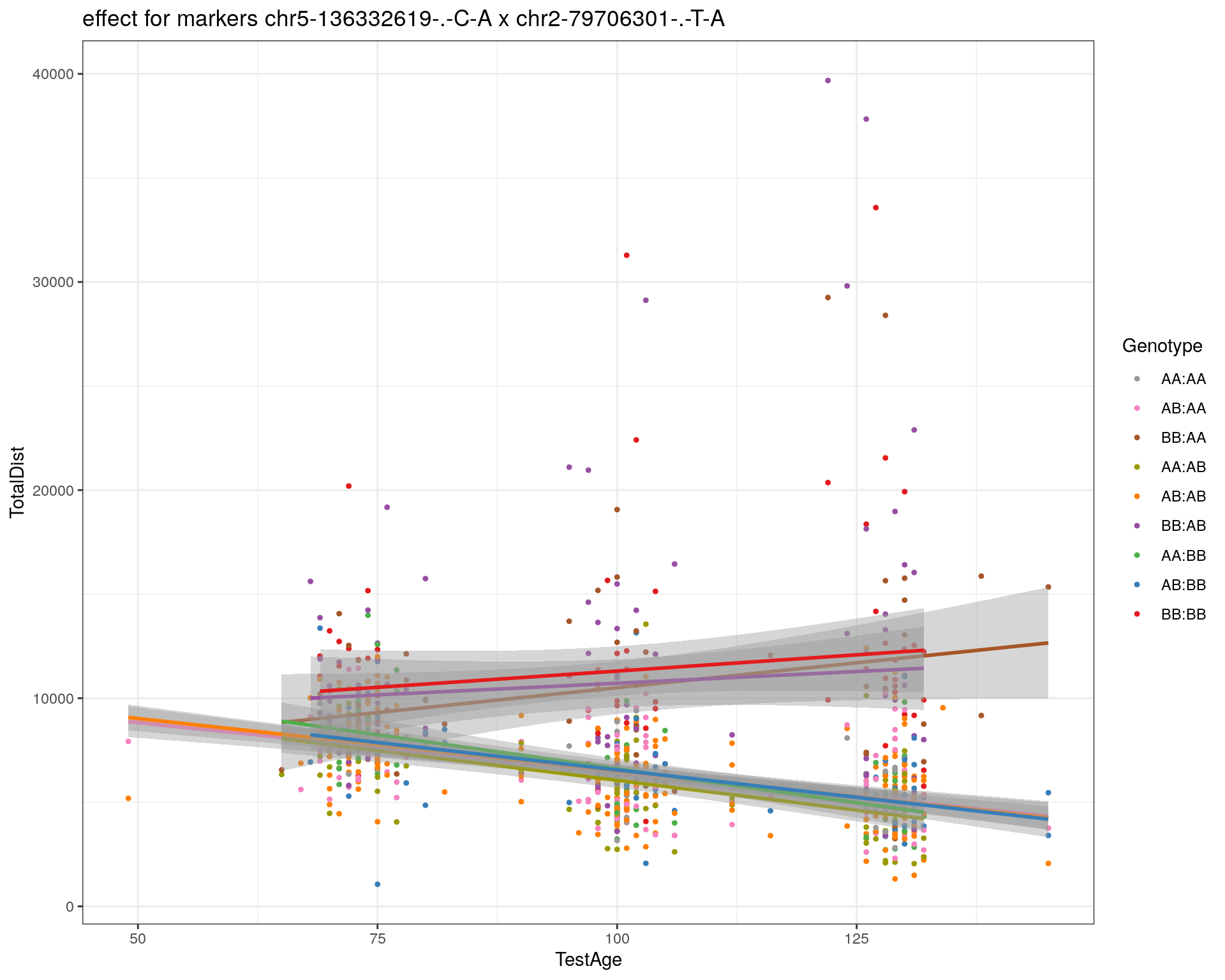
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
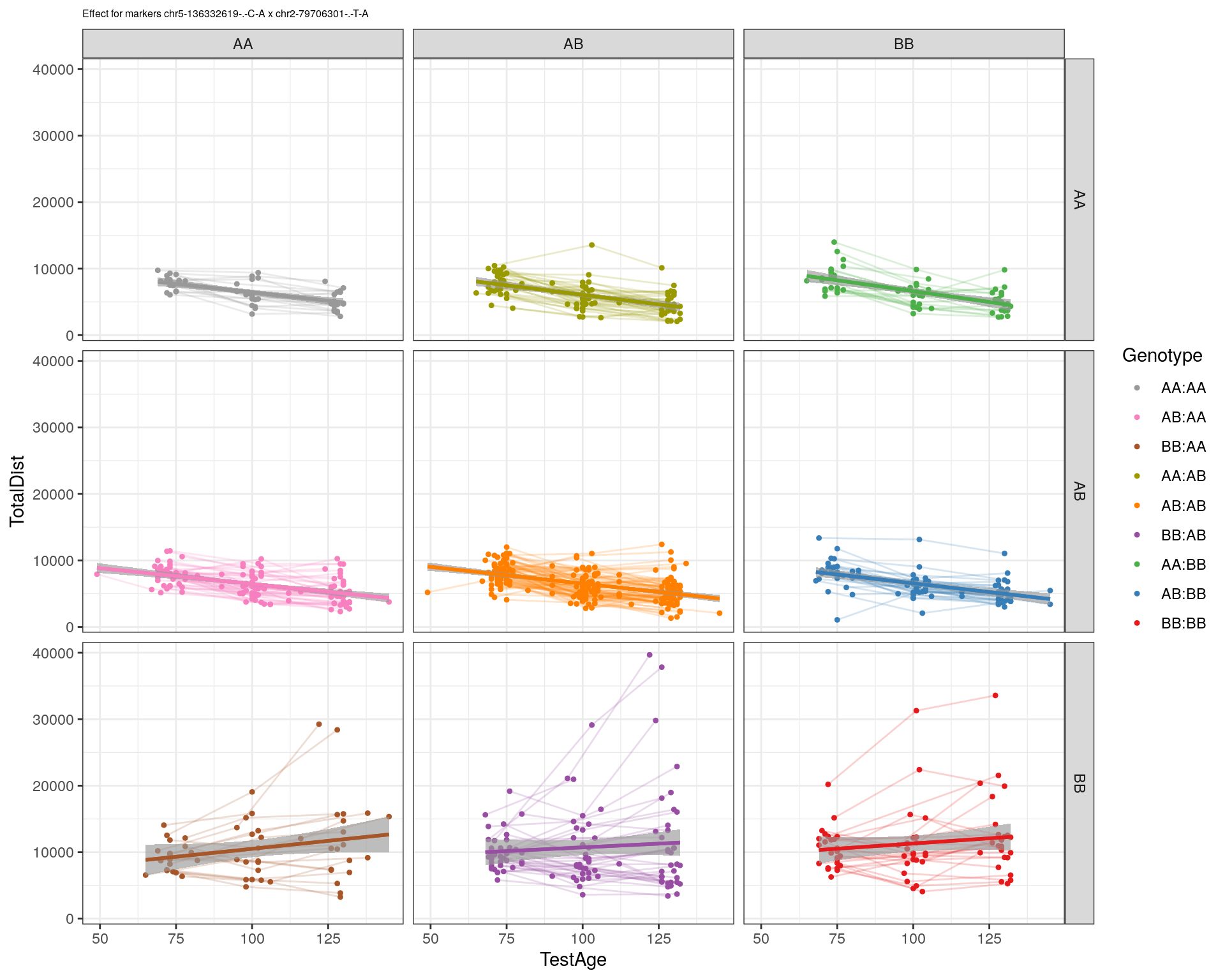
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr2-92367783-.-T-C"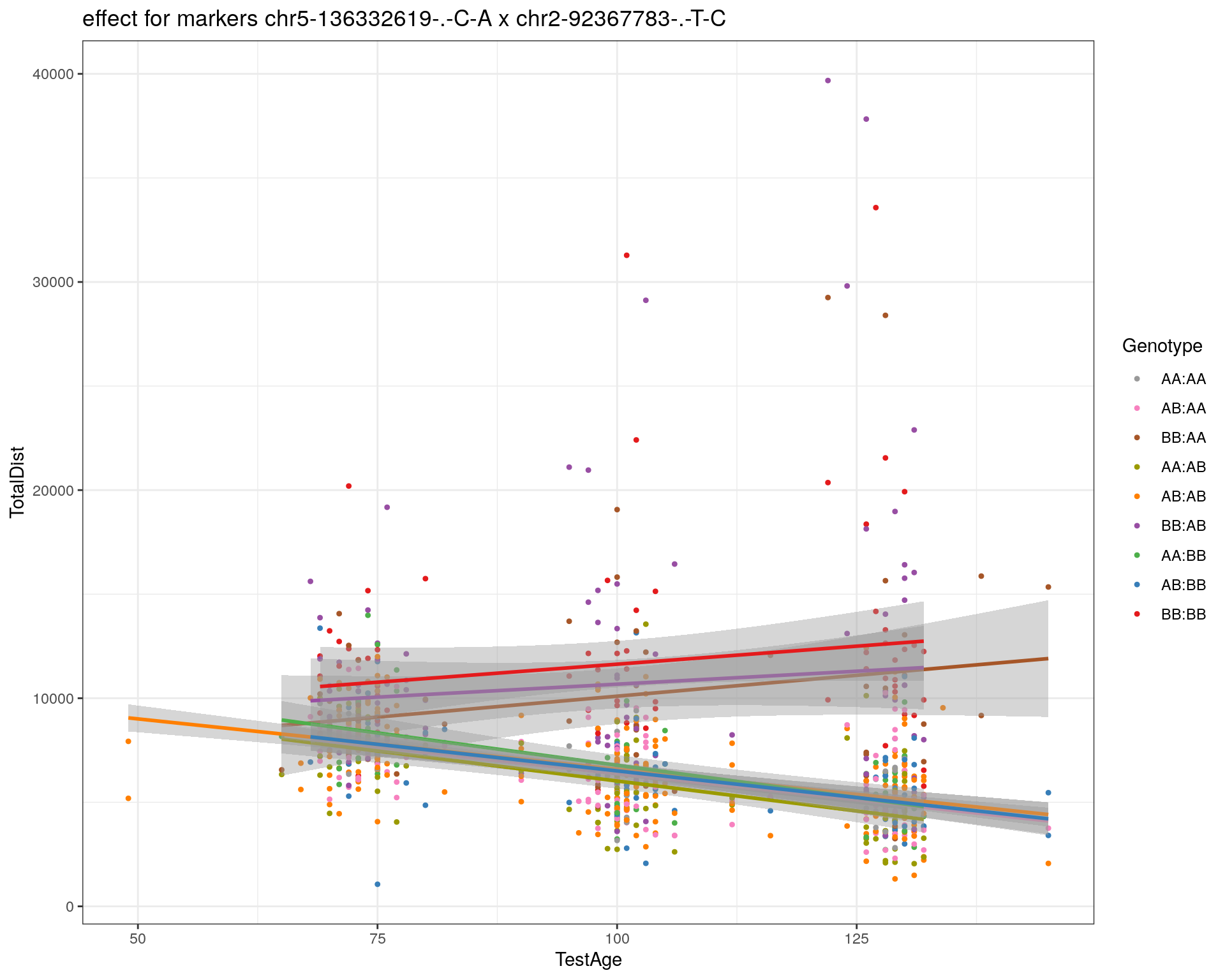
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
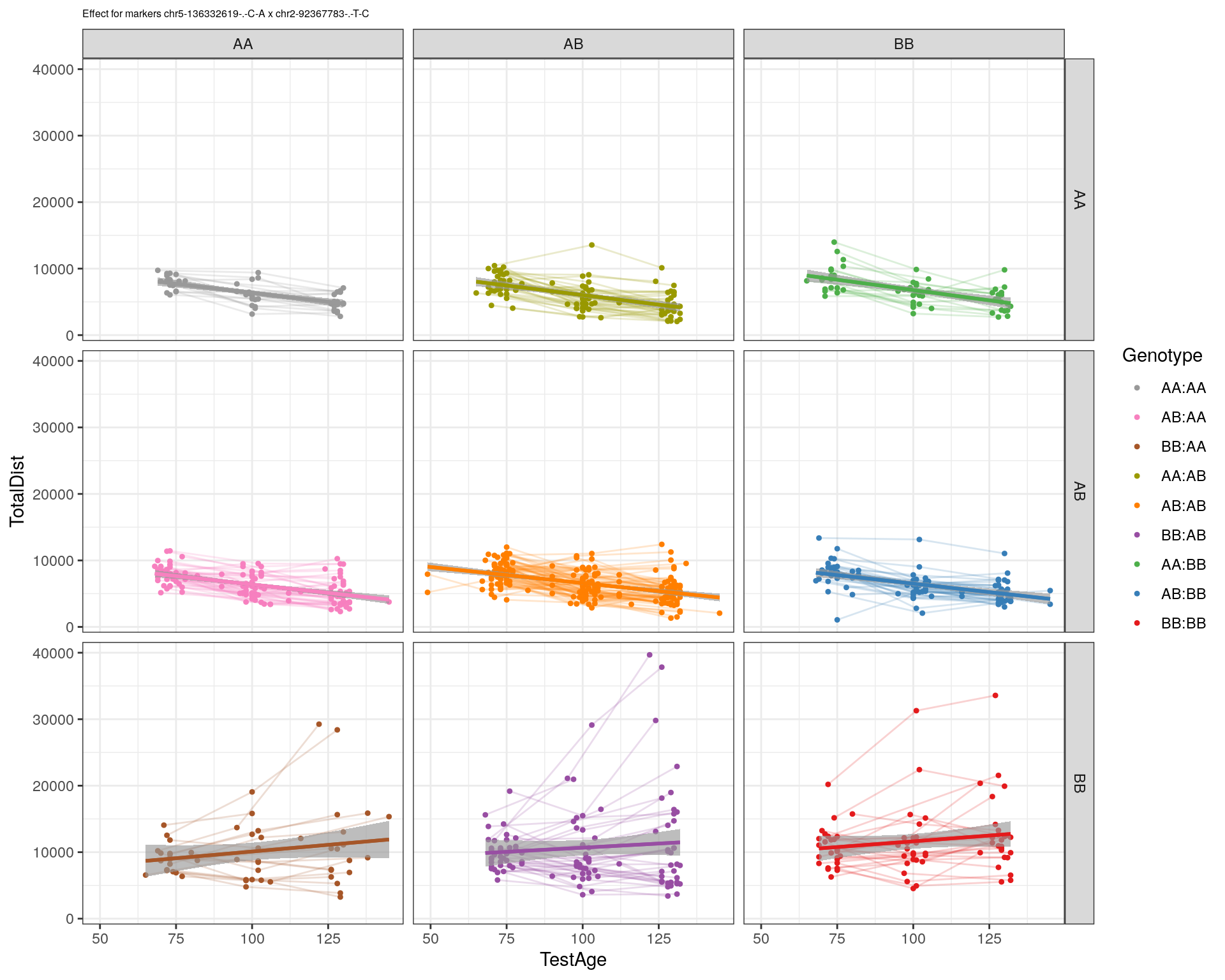
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr2-95118744-.-T-A"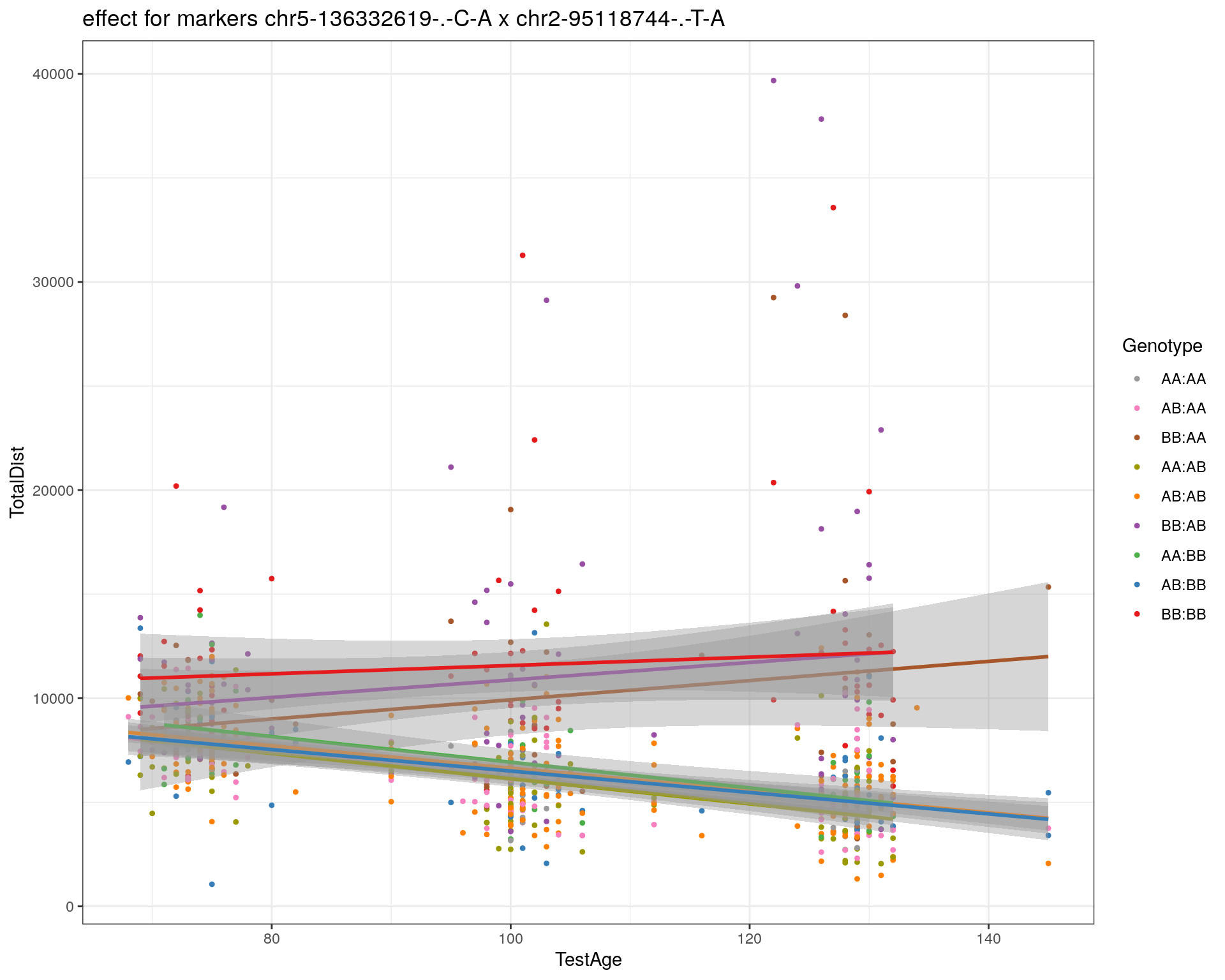
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
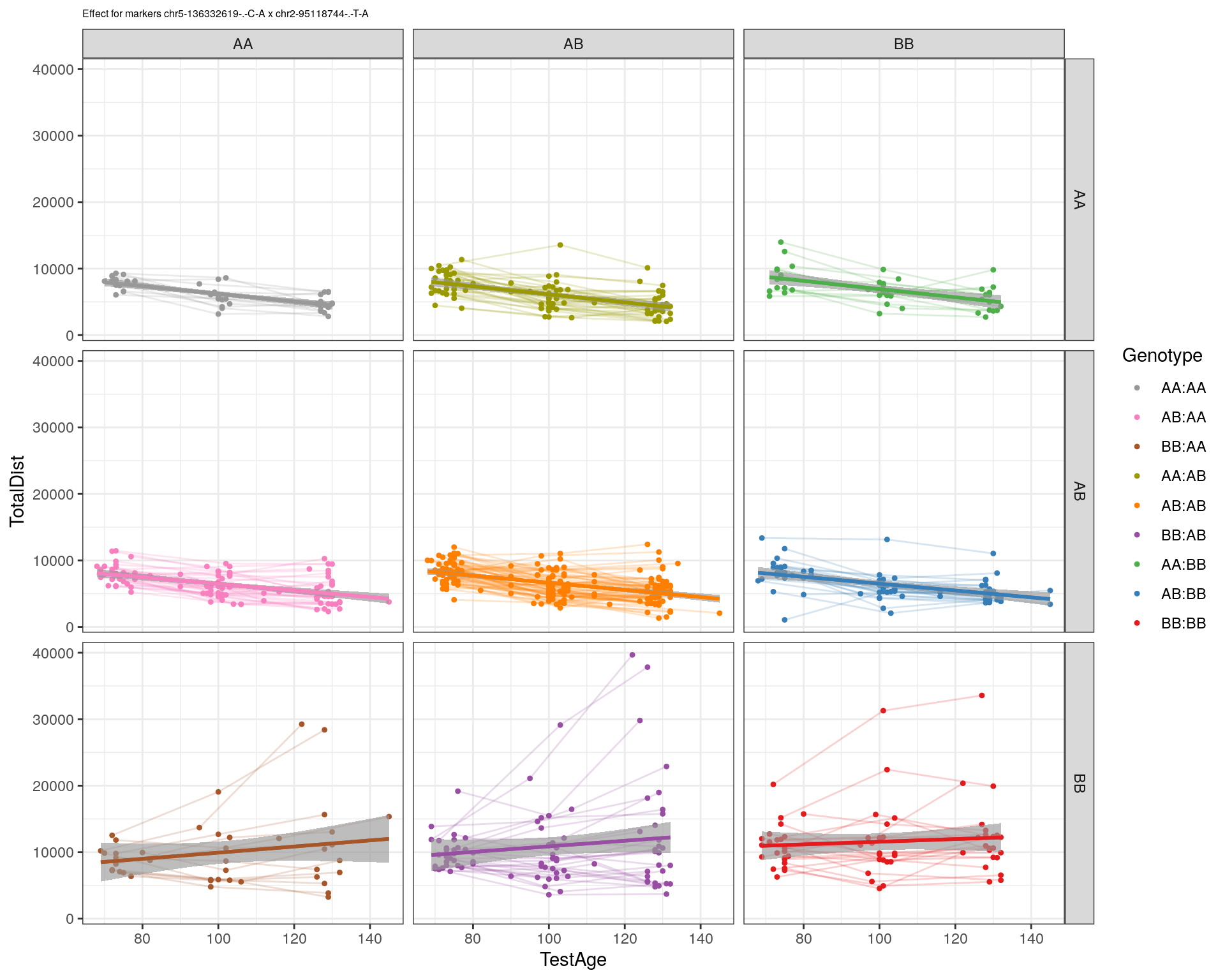
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr2-102169266-.-C-T"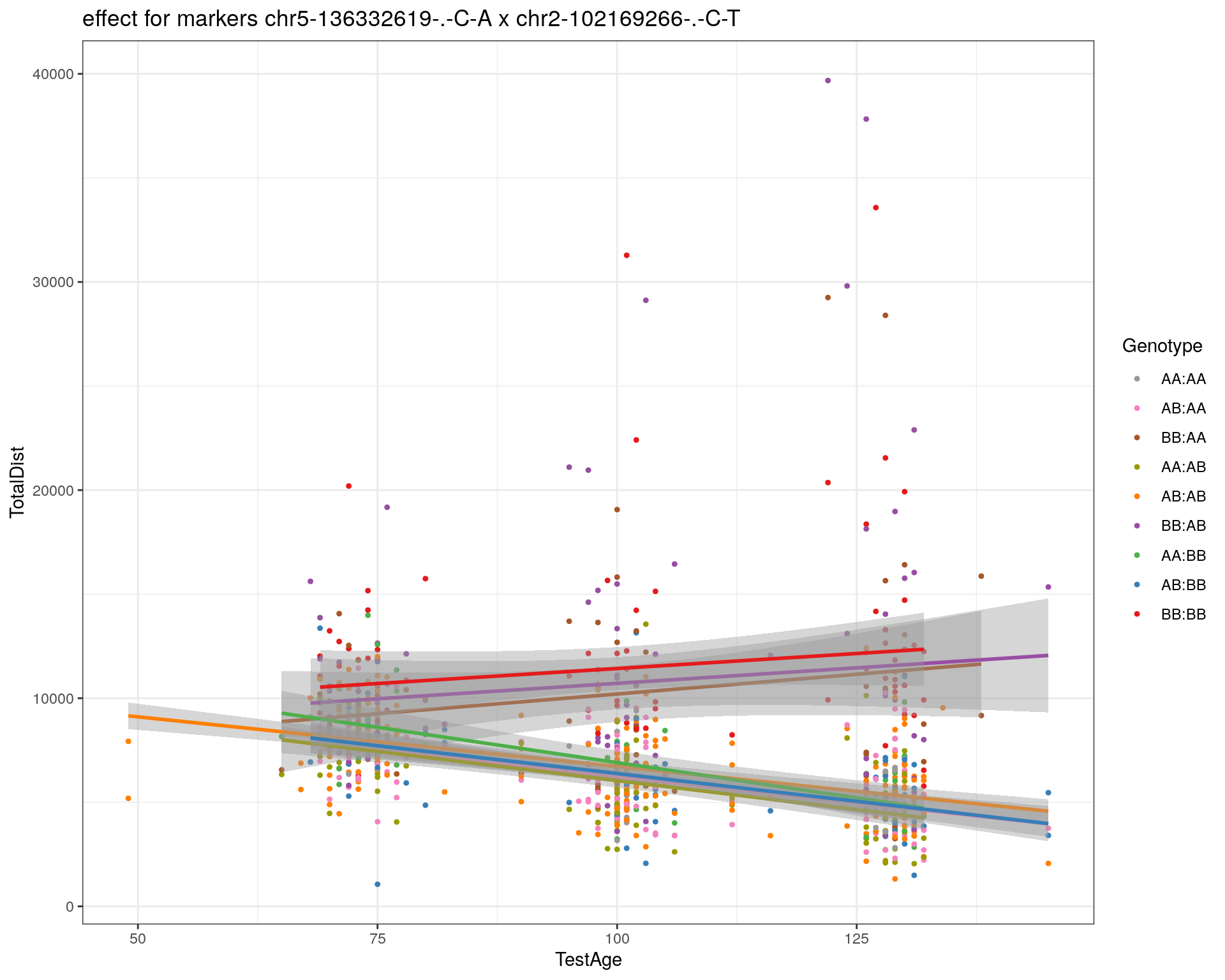
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
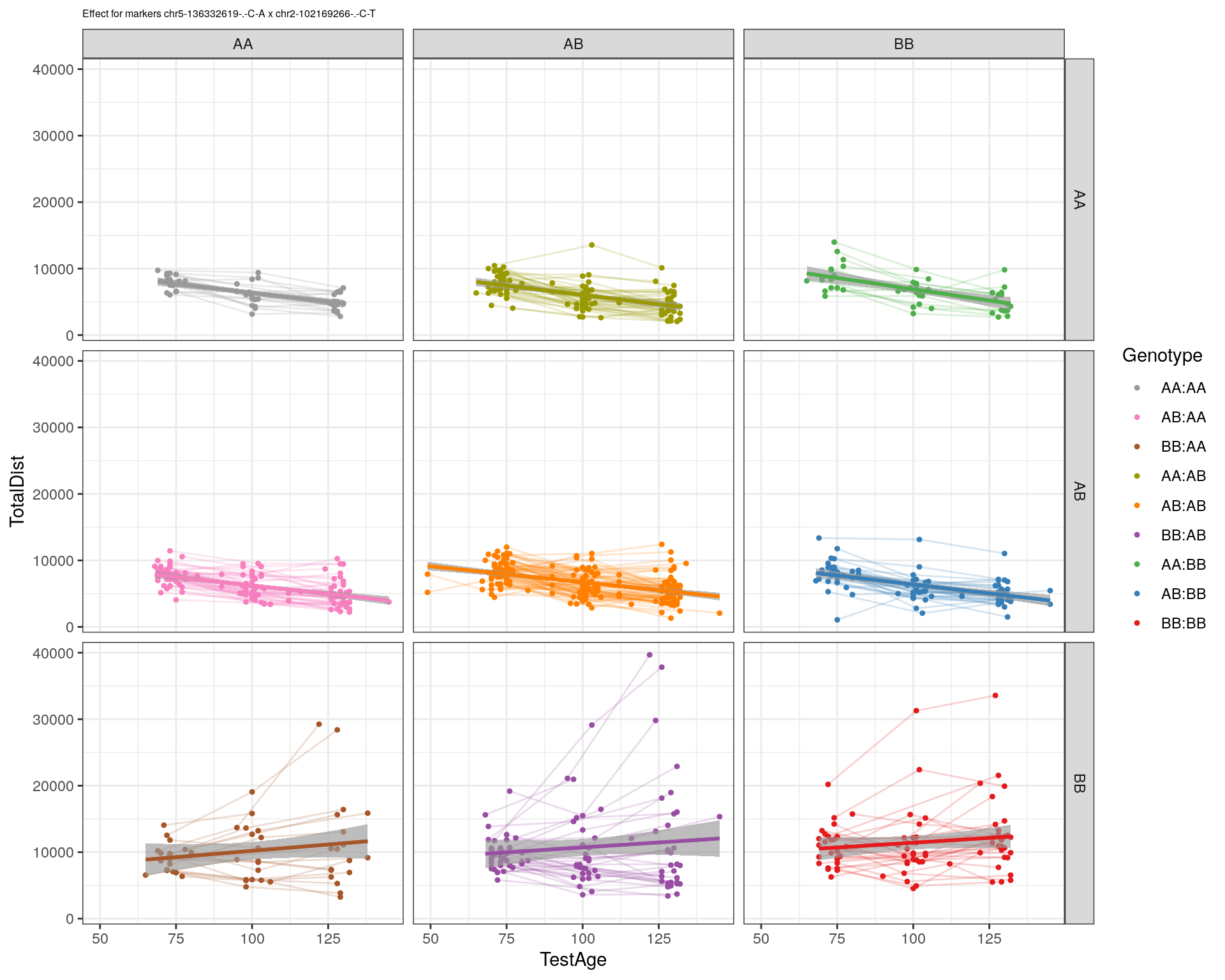
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
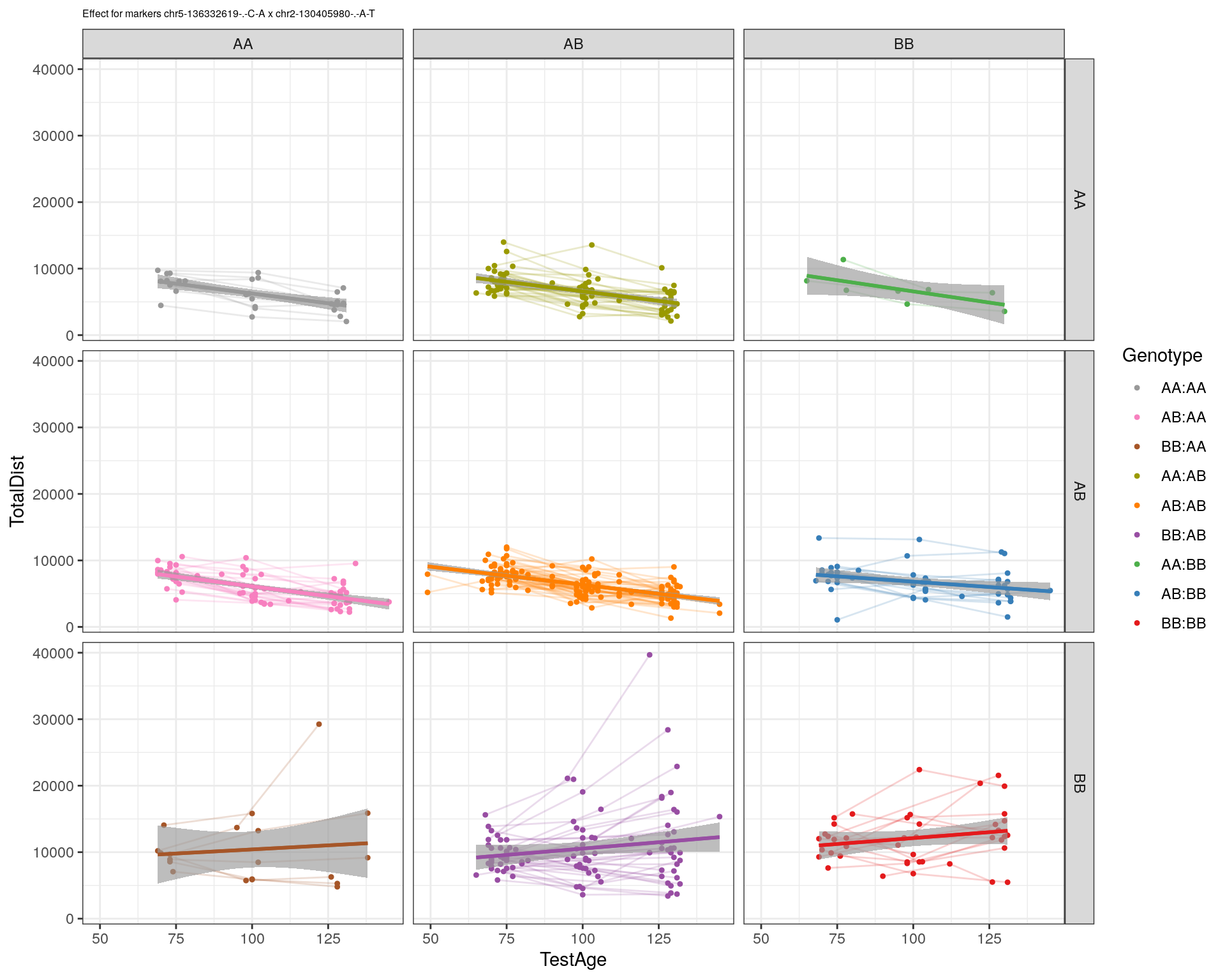
[1] "chr2-130405980-.-A-T"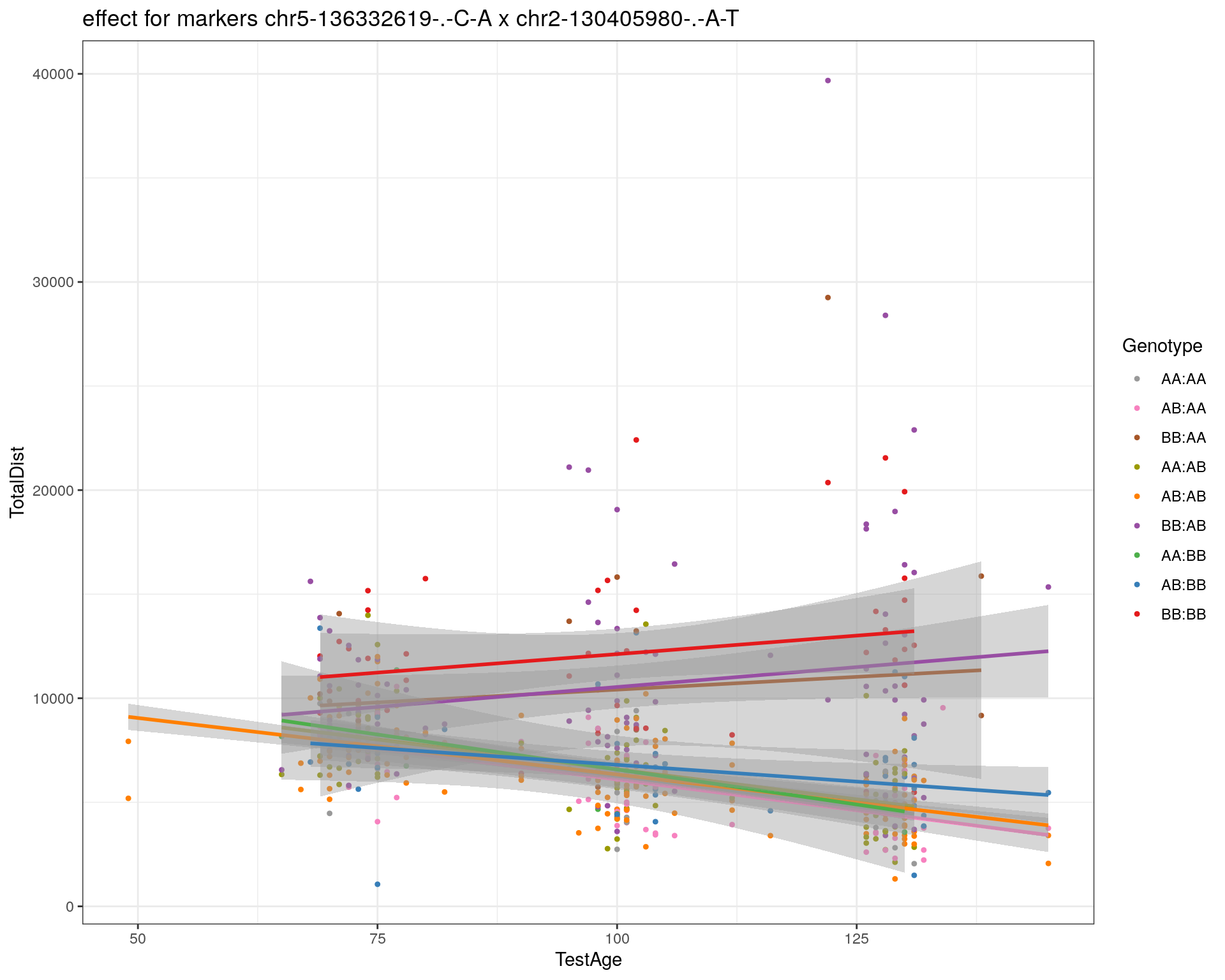
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
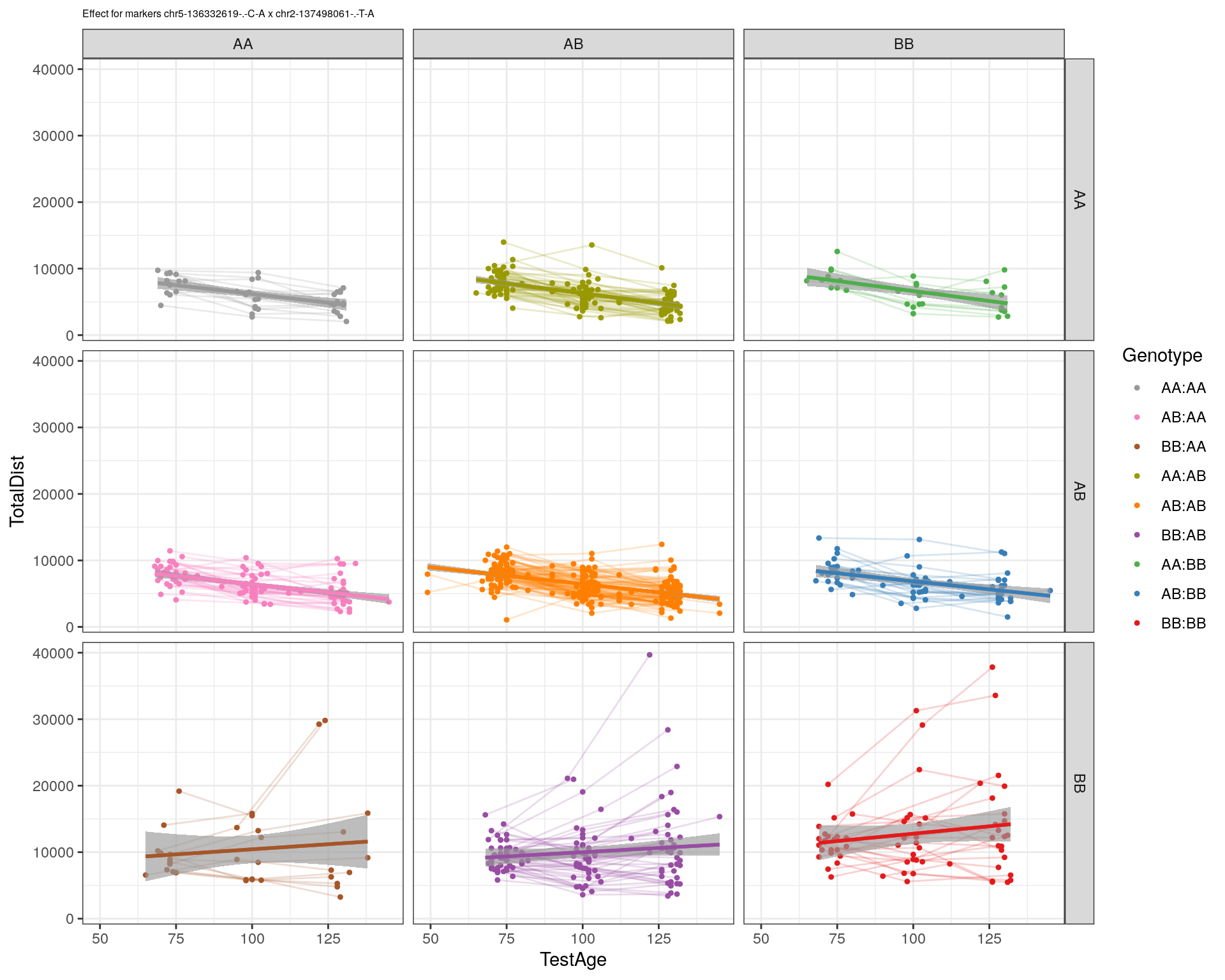
[1] "chr2-137498061-.-T-A"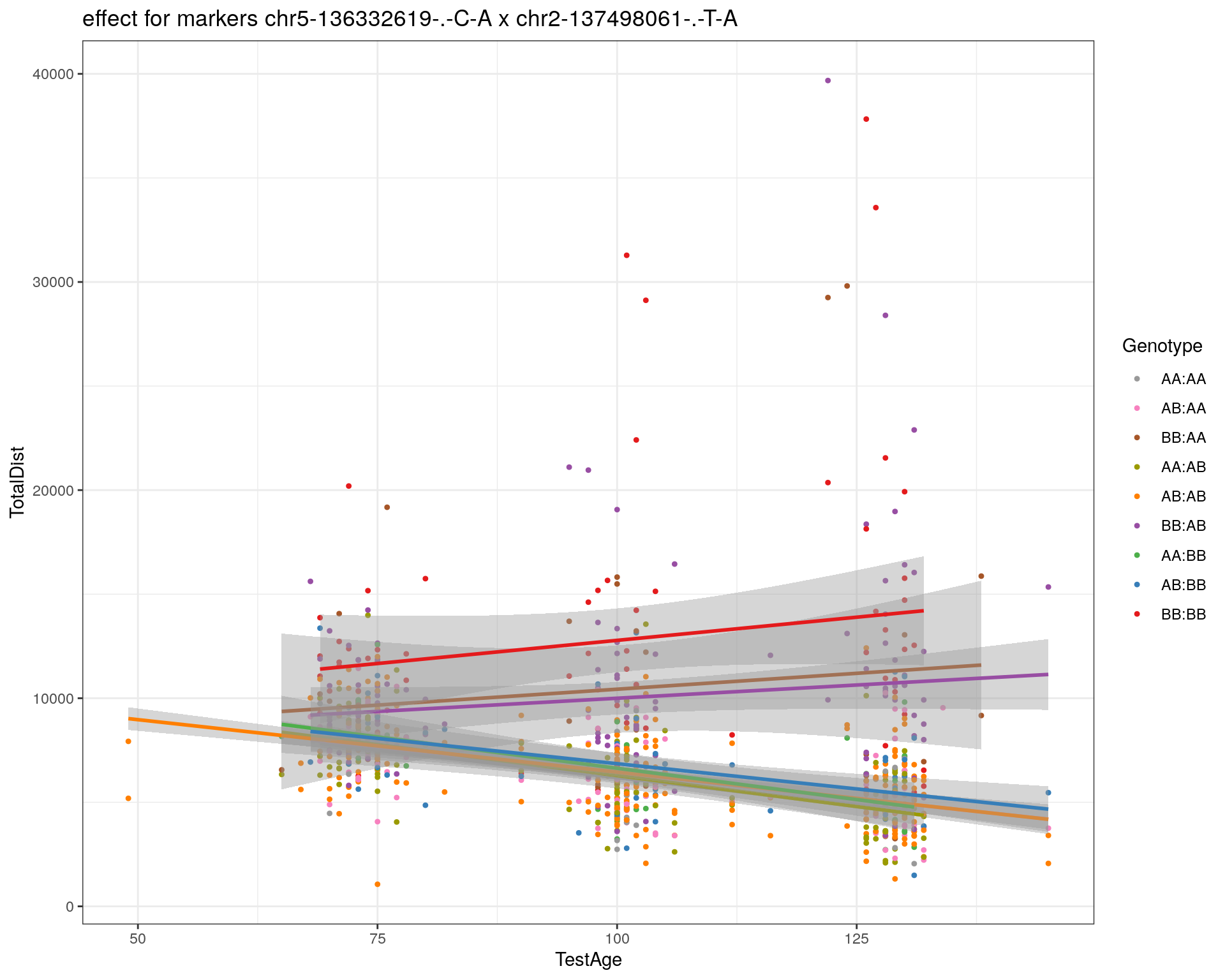
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr2-145128463-.-A-T"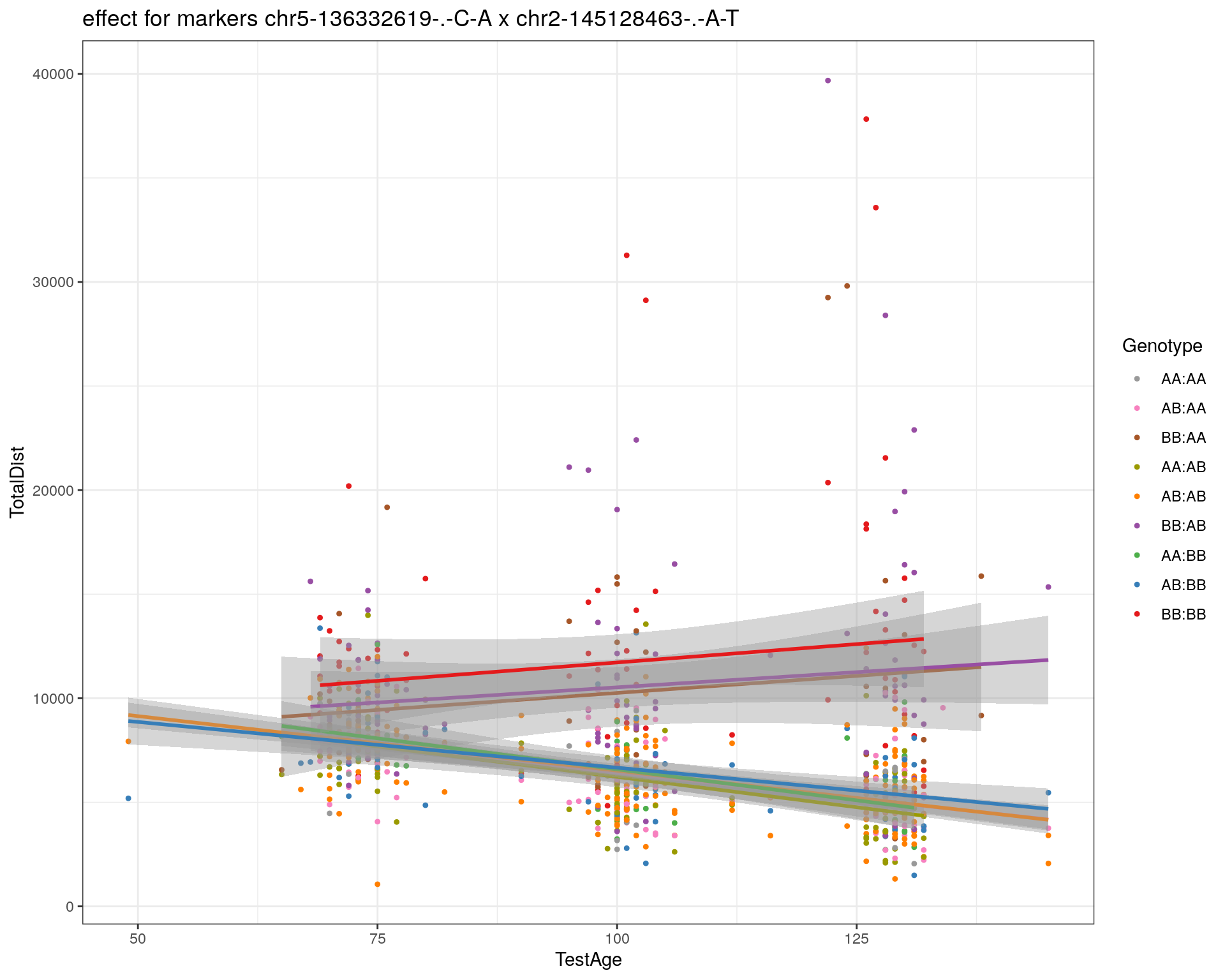
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-59218564-.-T-A"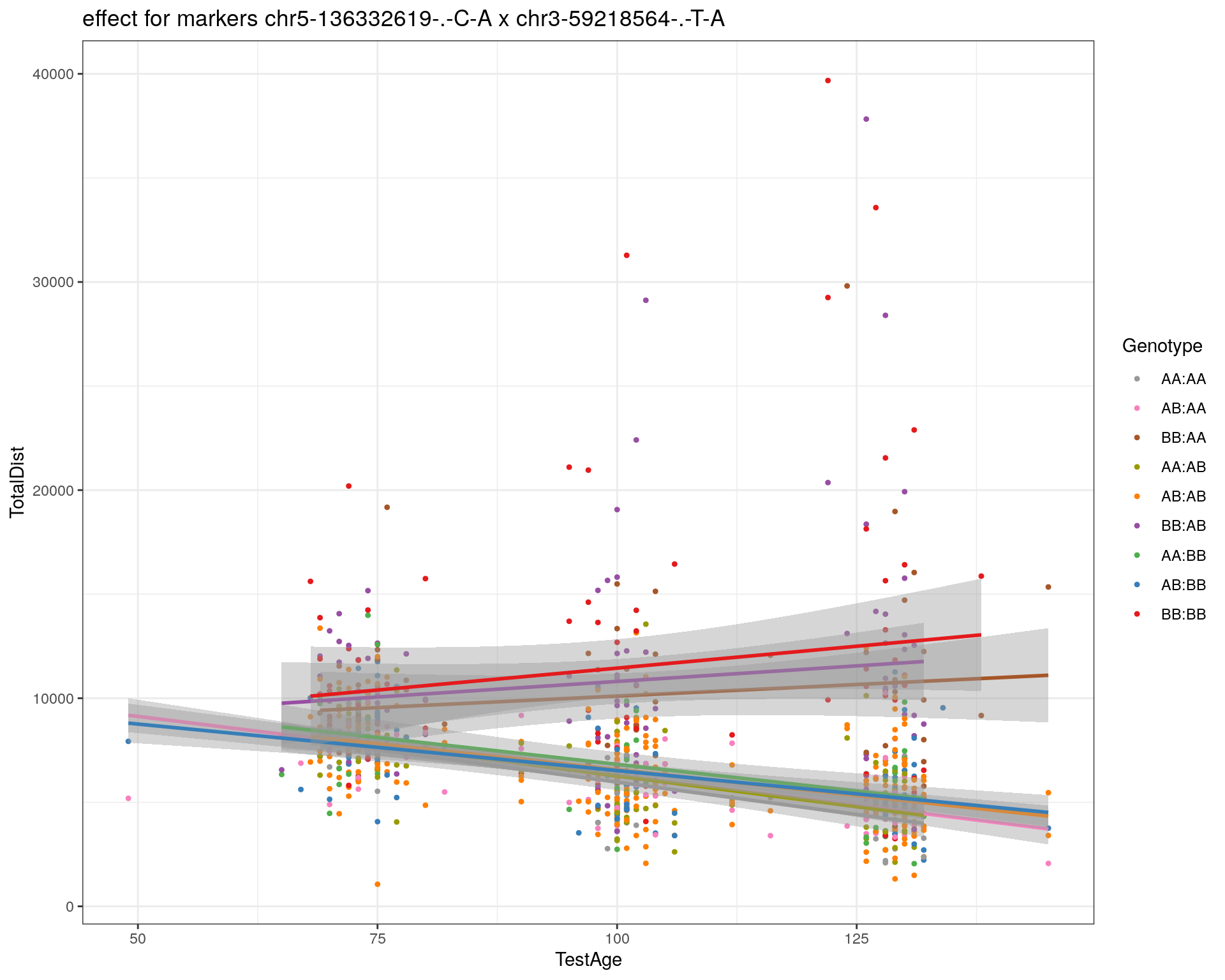
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-98881230-.-G-A"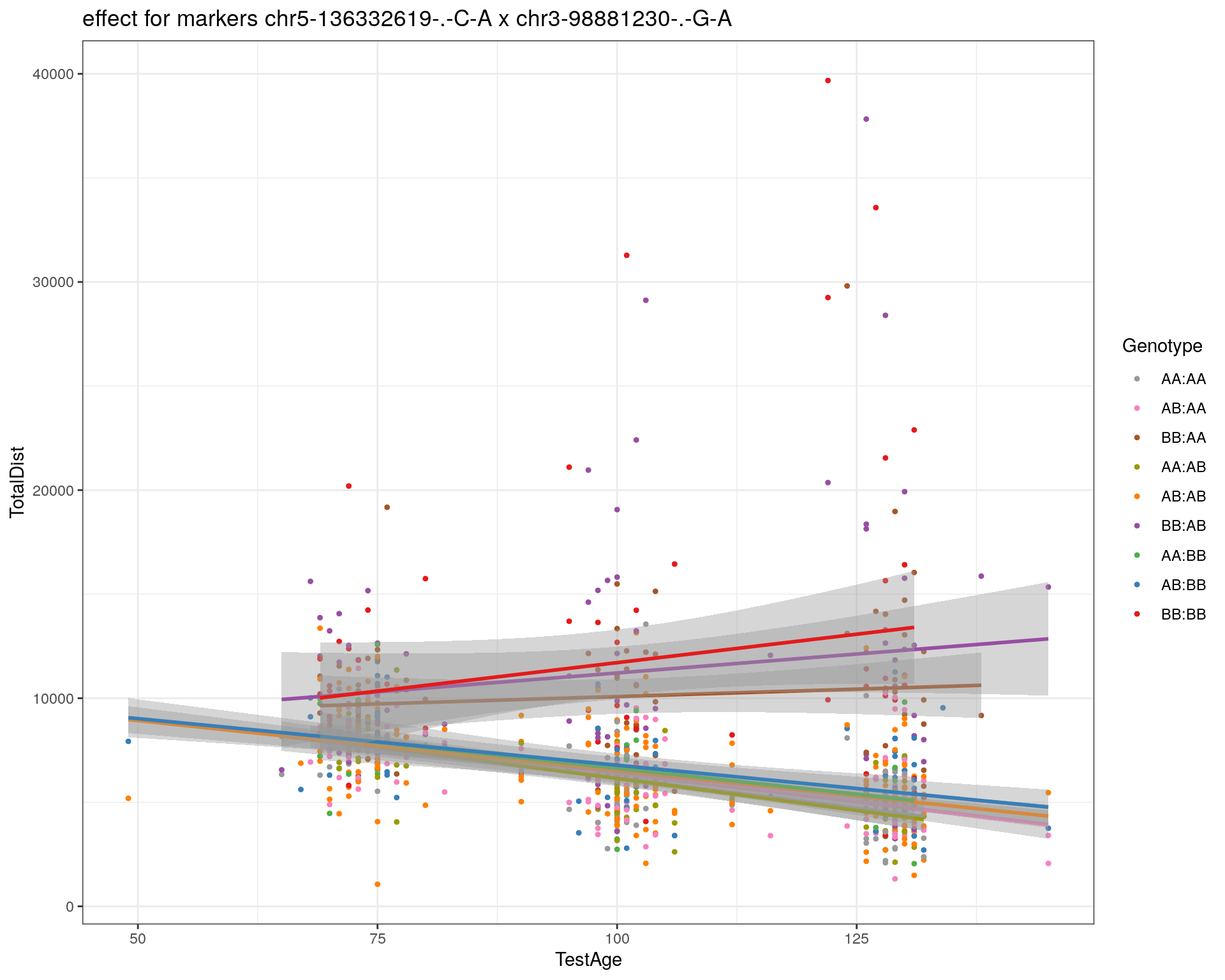
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
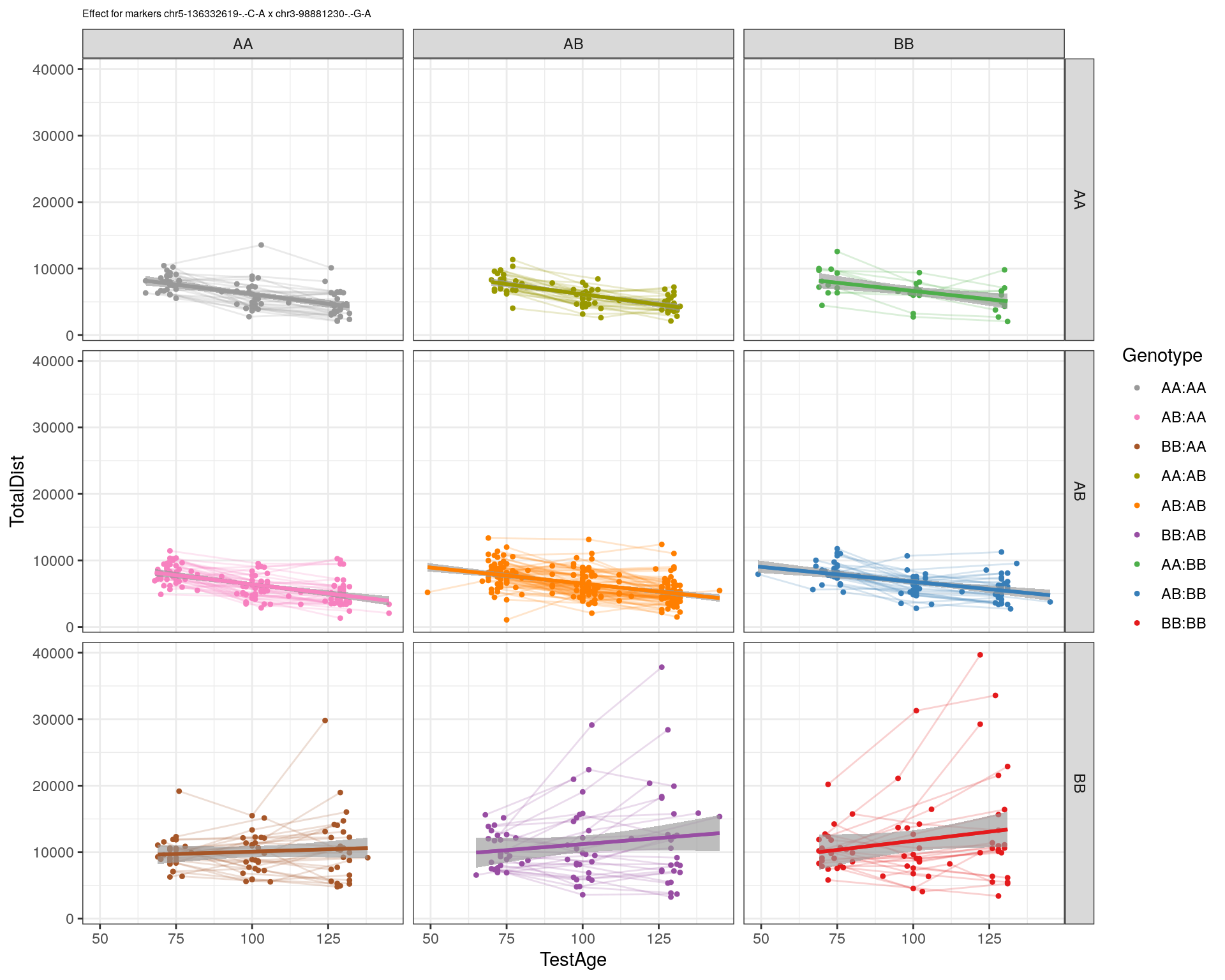
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-106985708-.-A-C"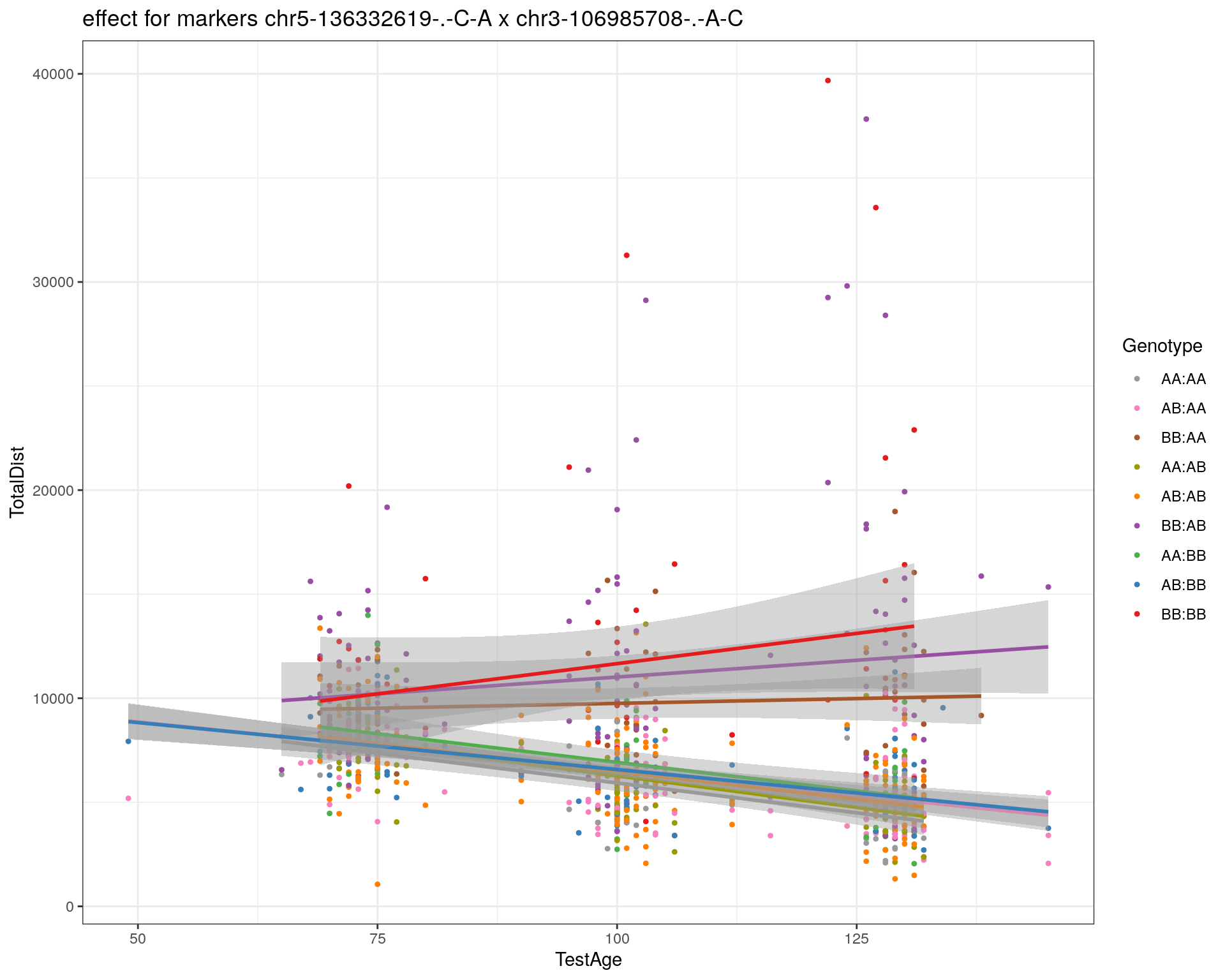
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
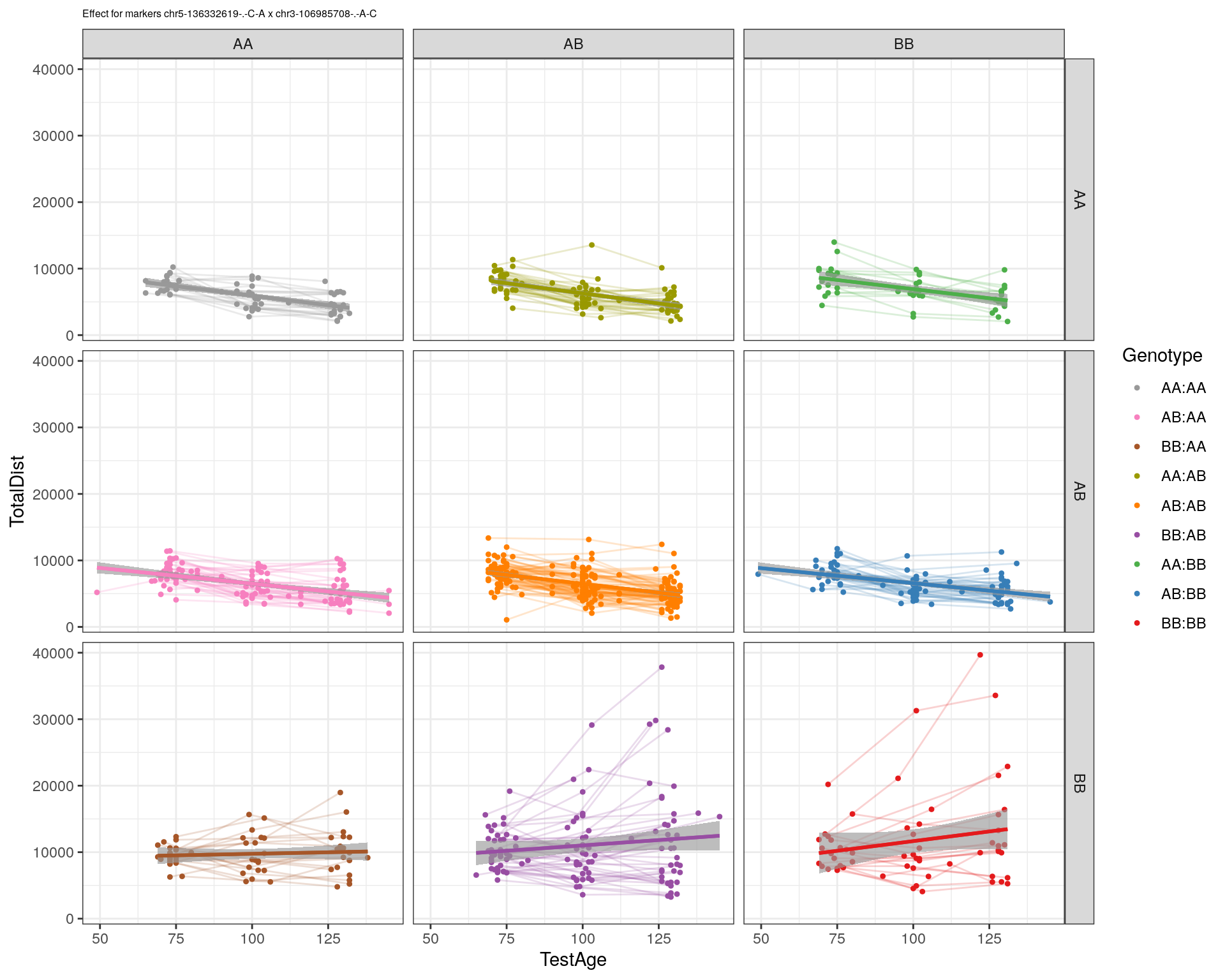
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-135614002-.-T-G"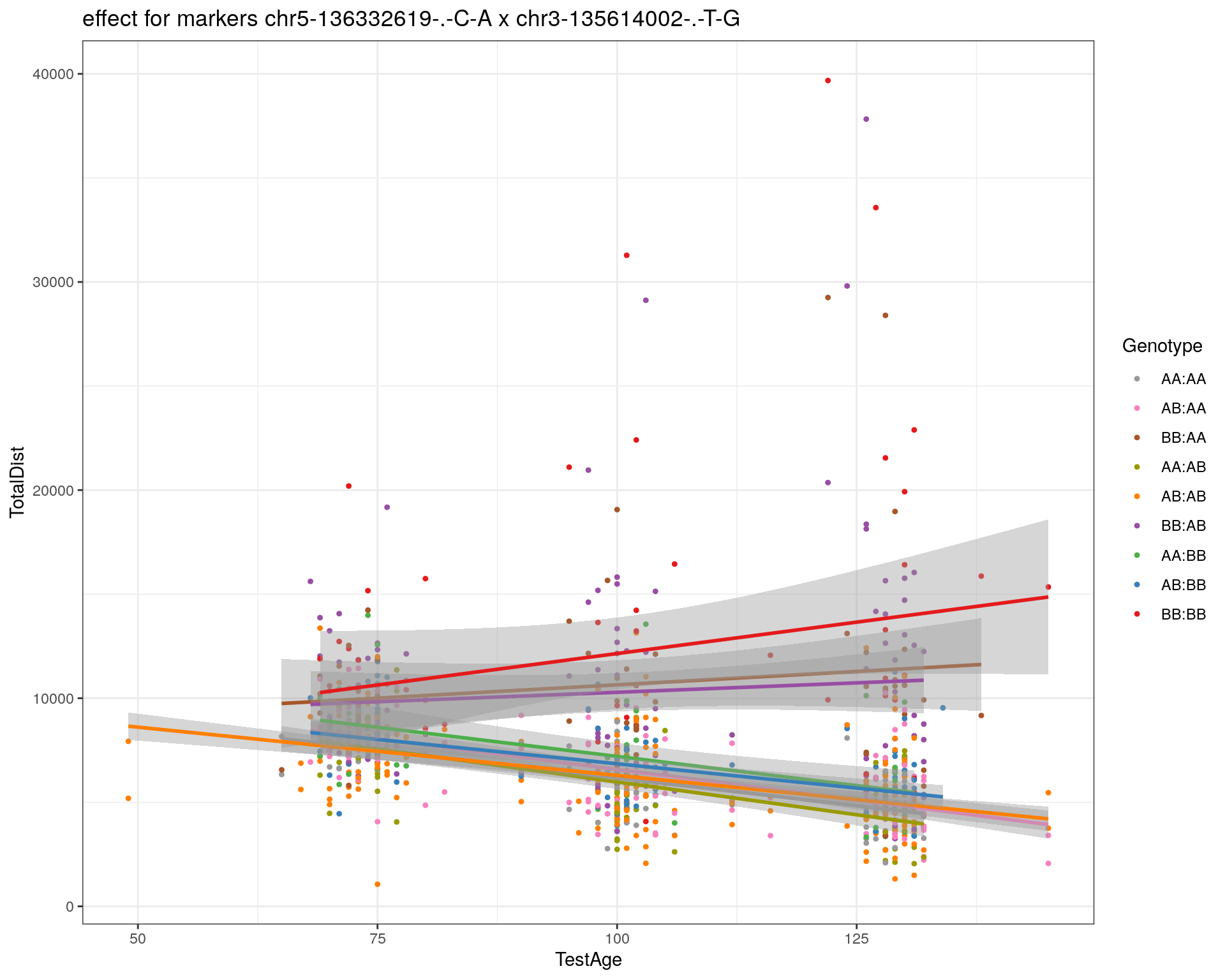
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
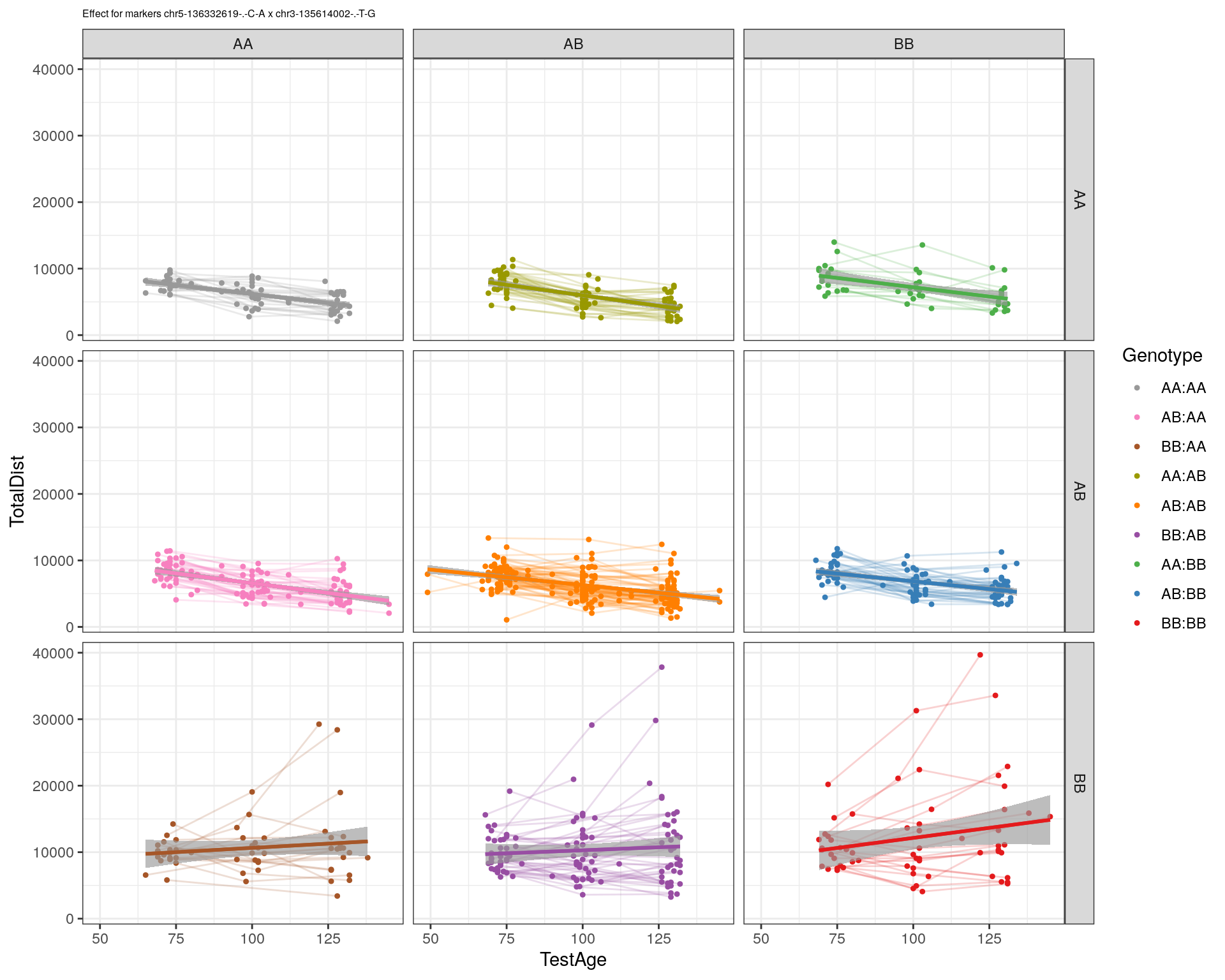
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-145286160-.-C-A"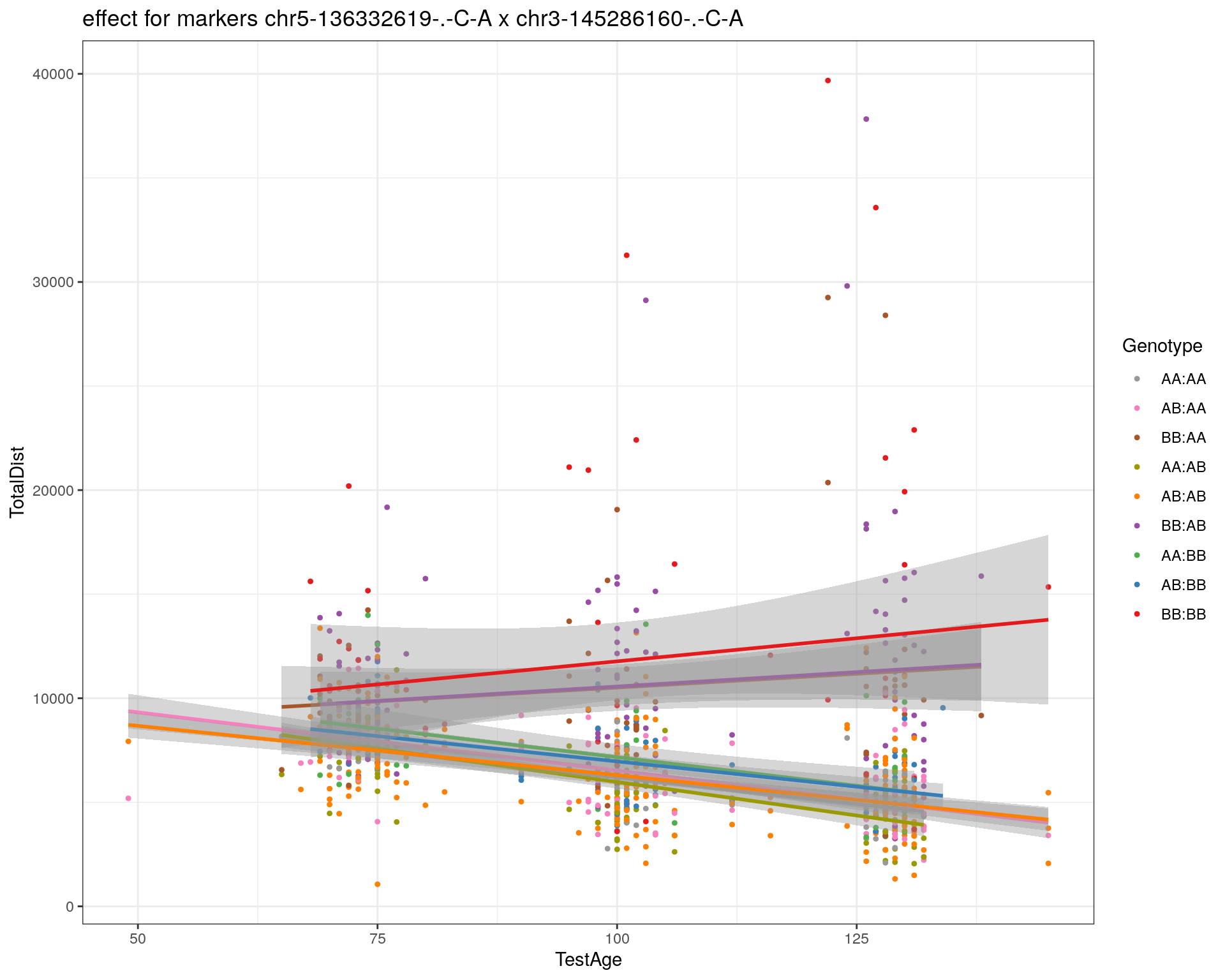
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
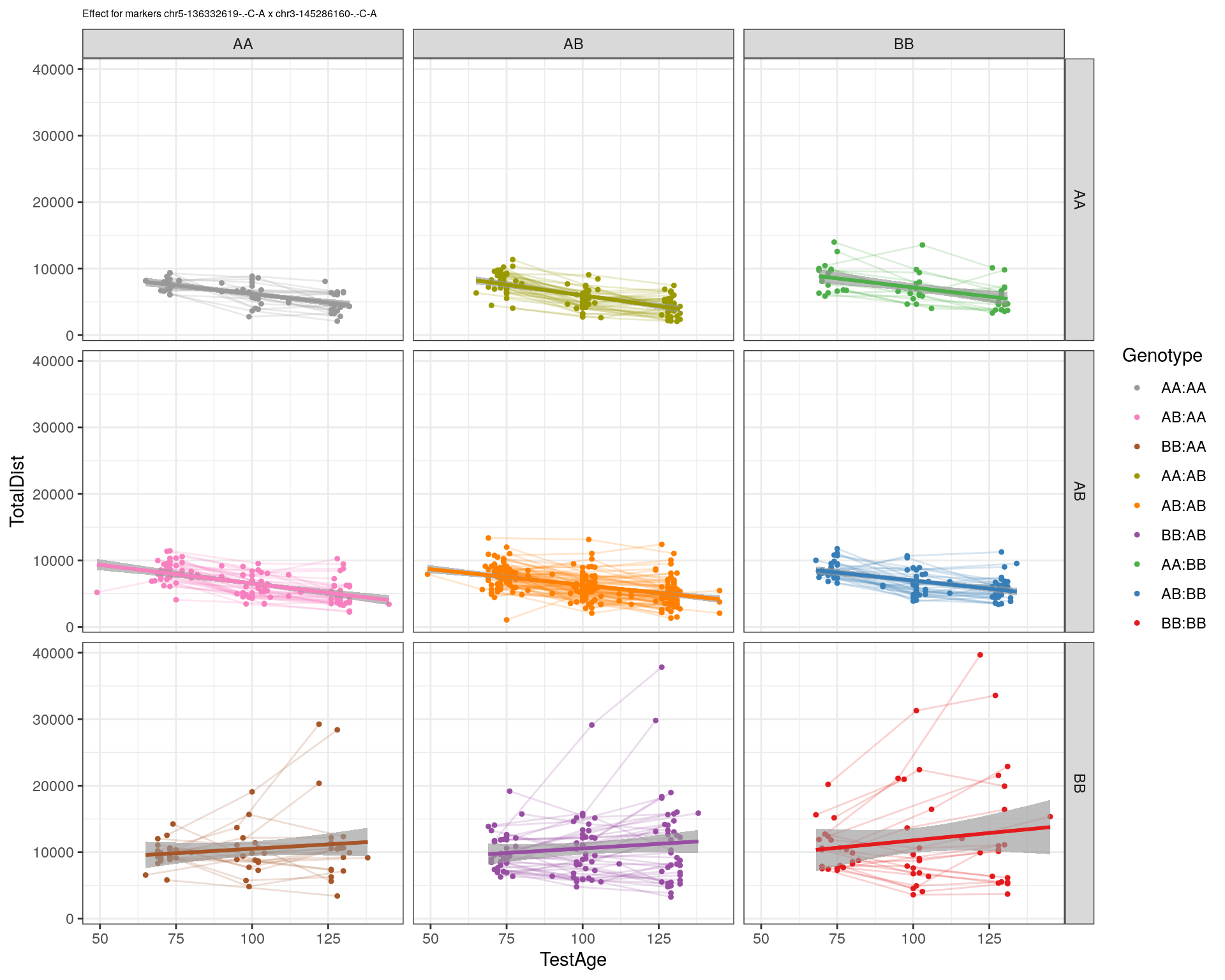
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-146750531-.-A-G"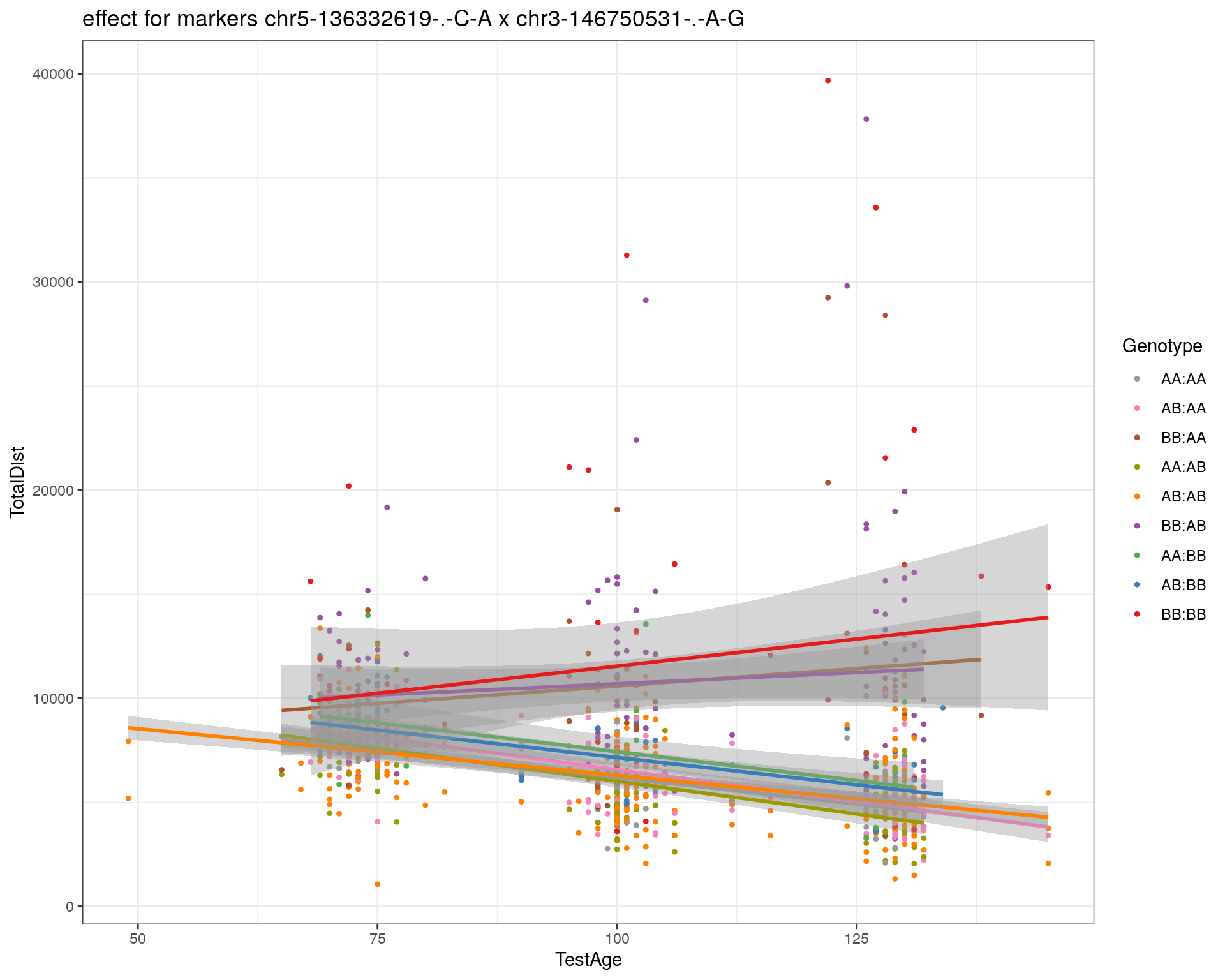
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
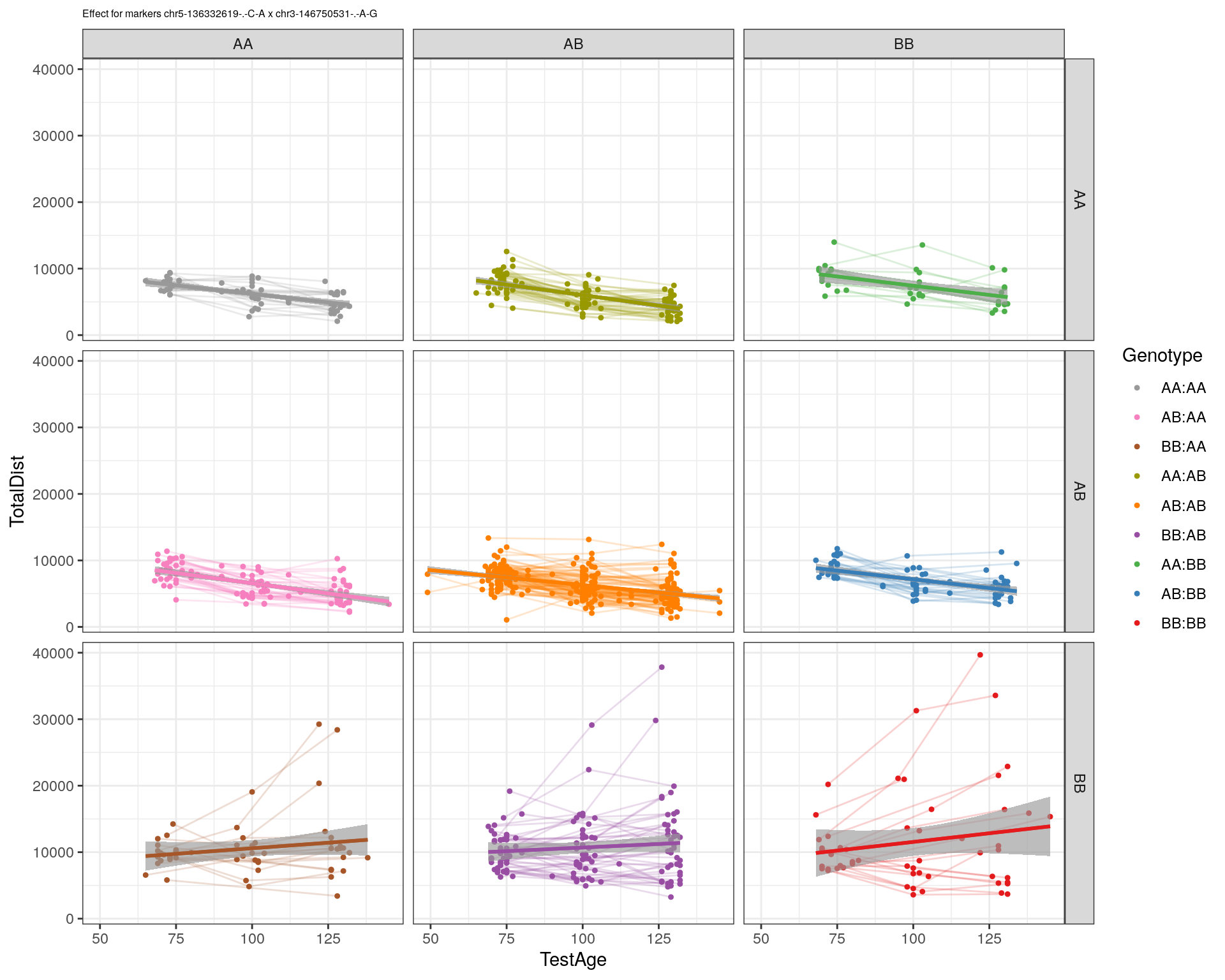
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr3-152699551-.-G-A"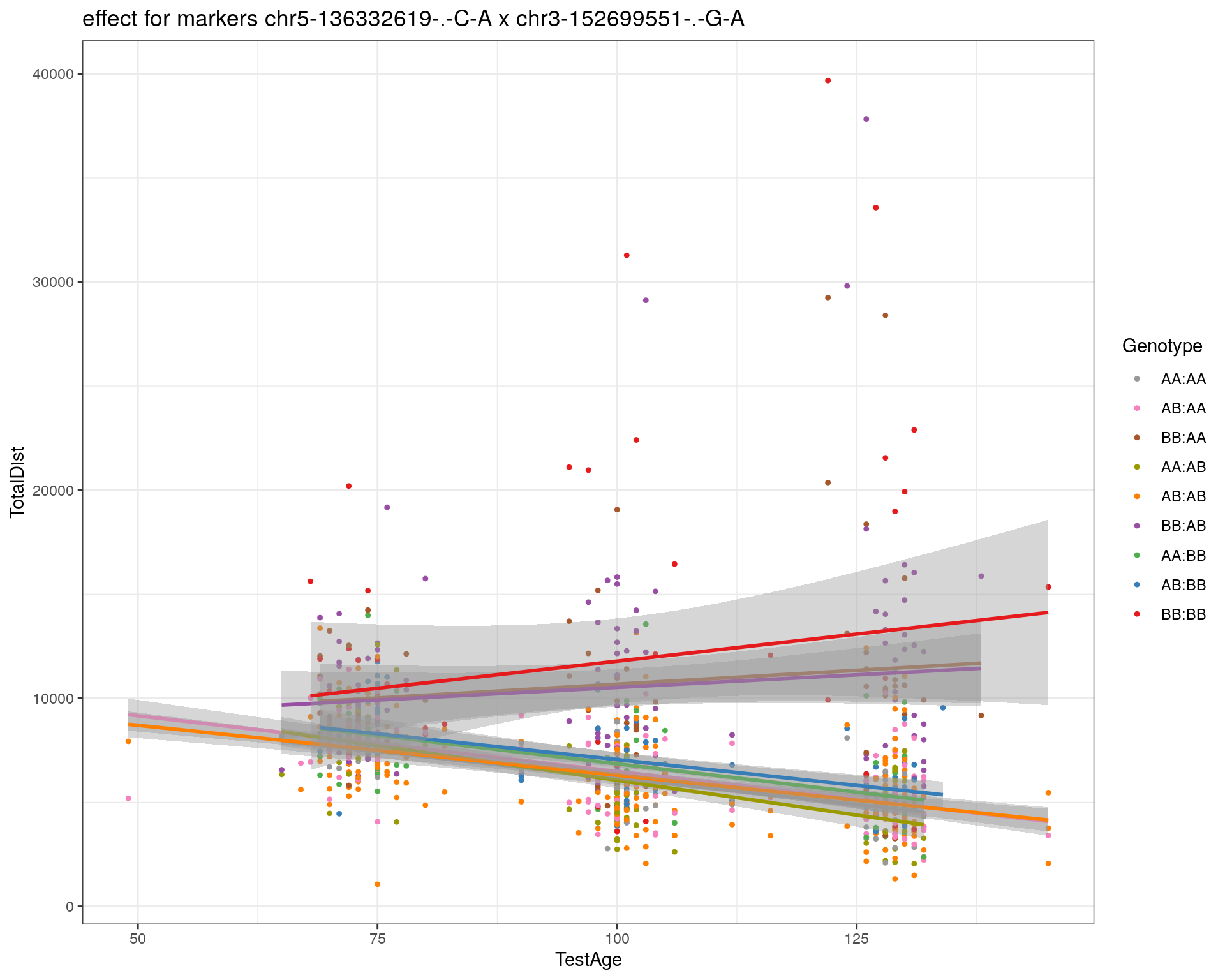
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
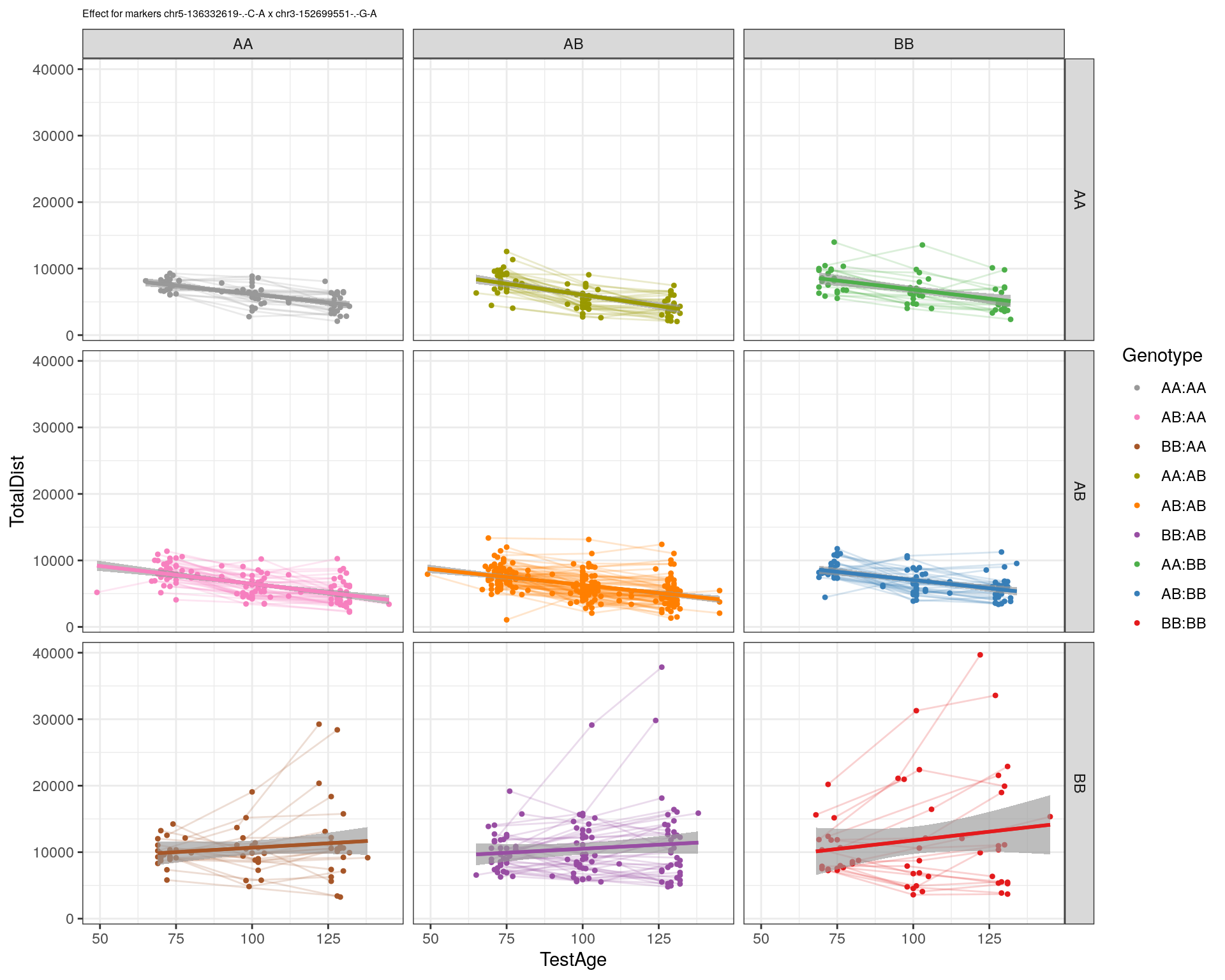
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr4-32821418-.-C-T"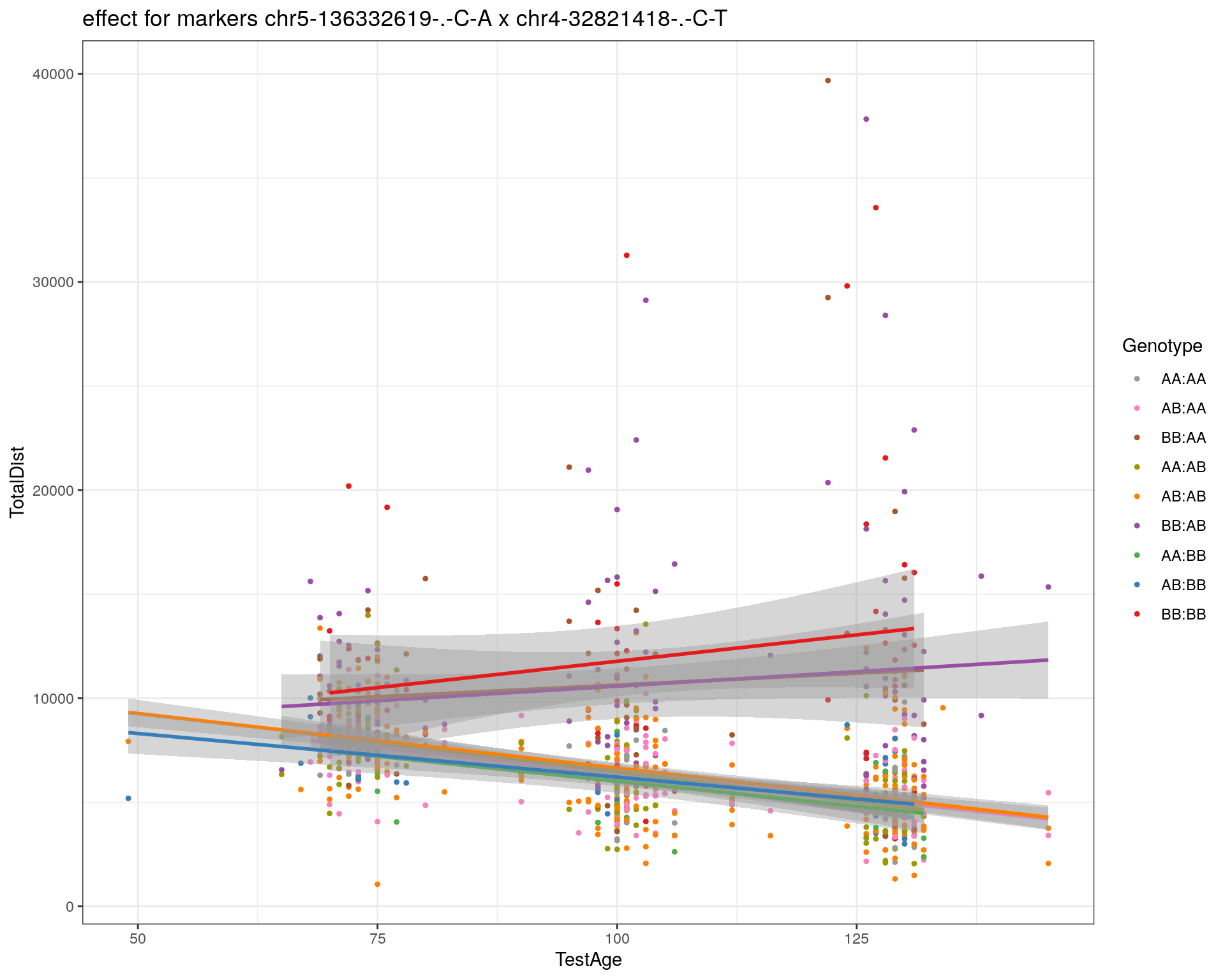
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
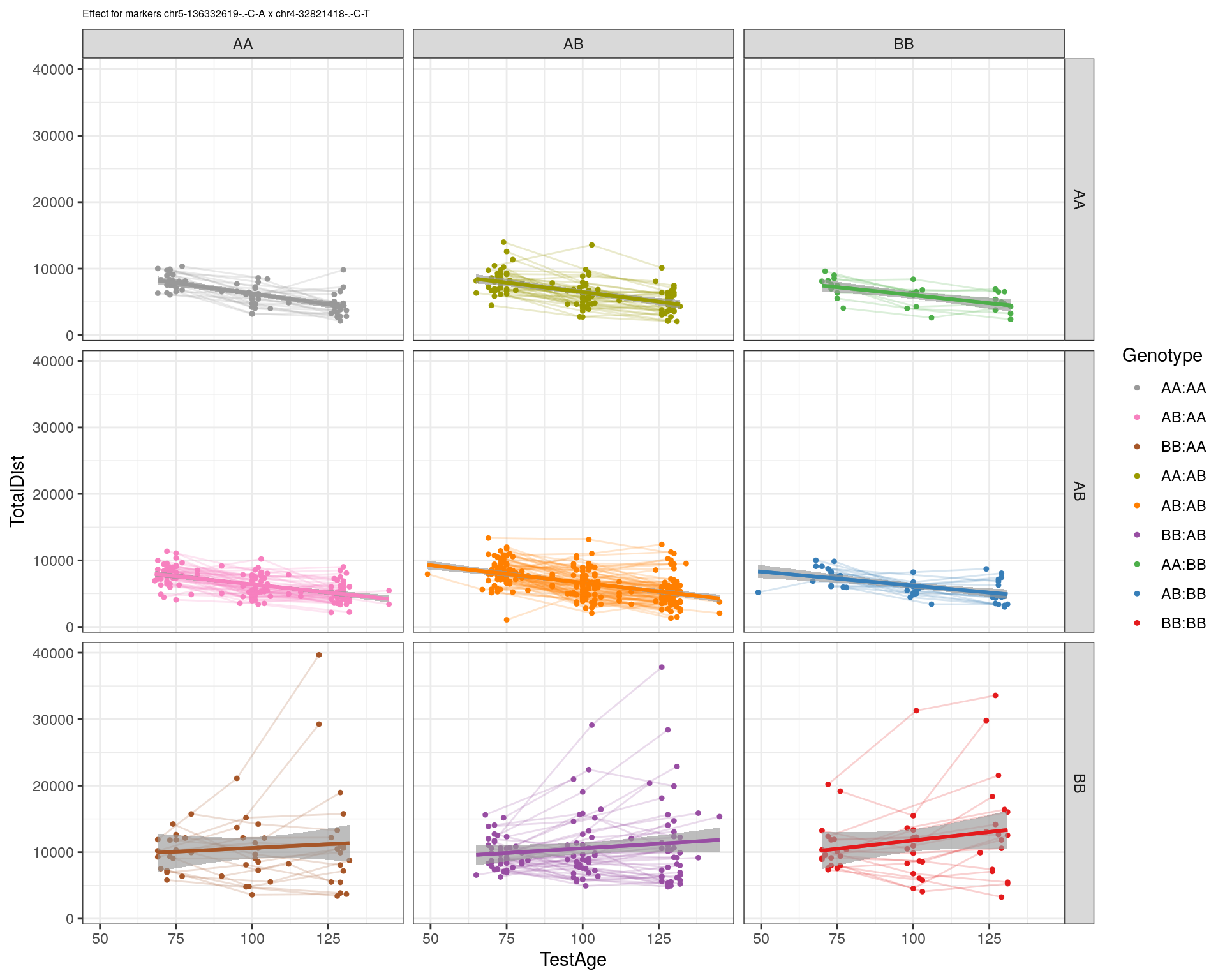
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-75264187-.-T-C"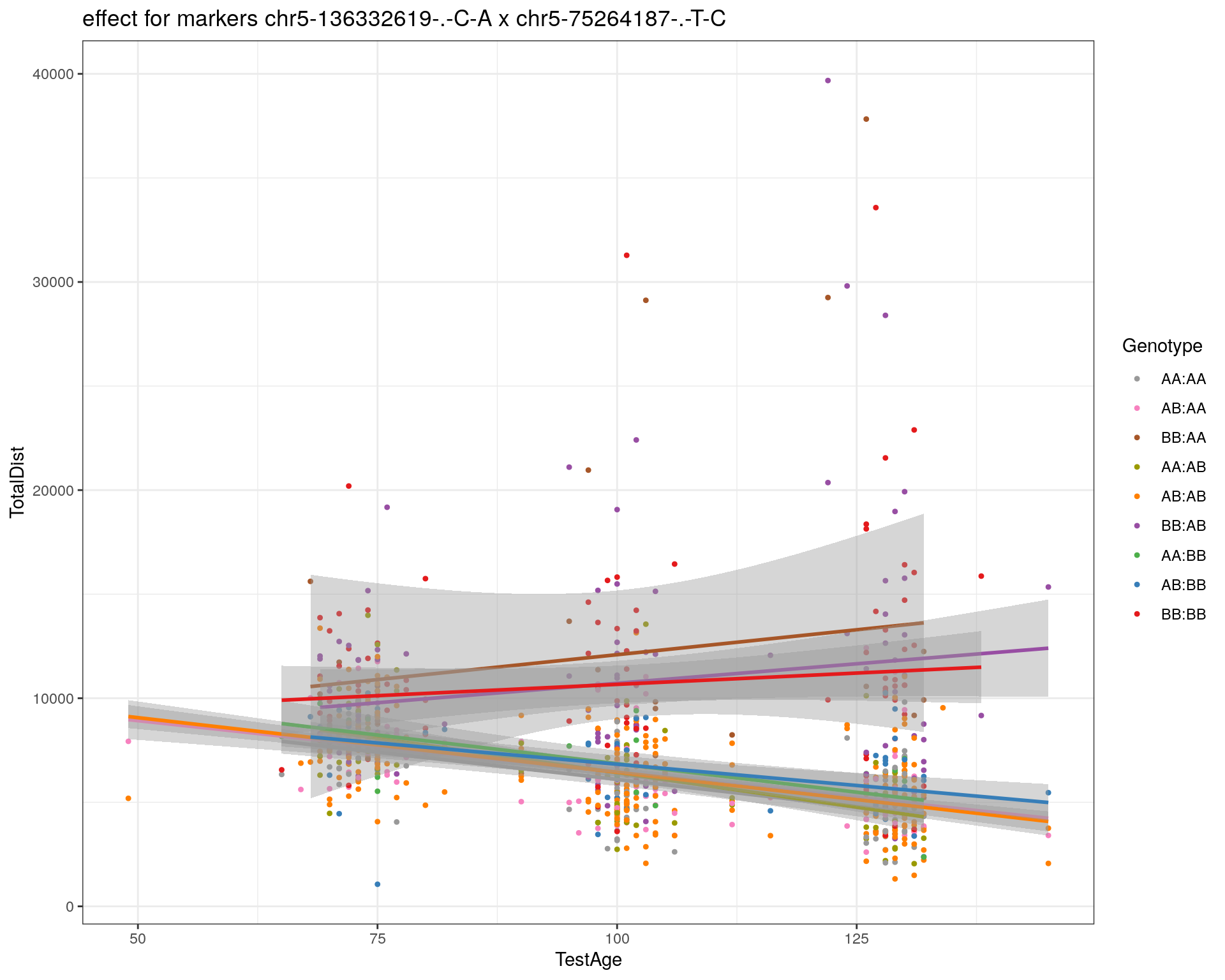
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
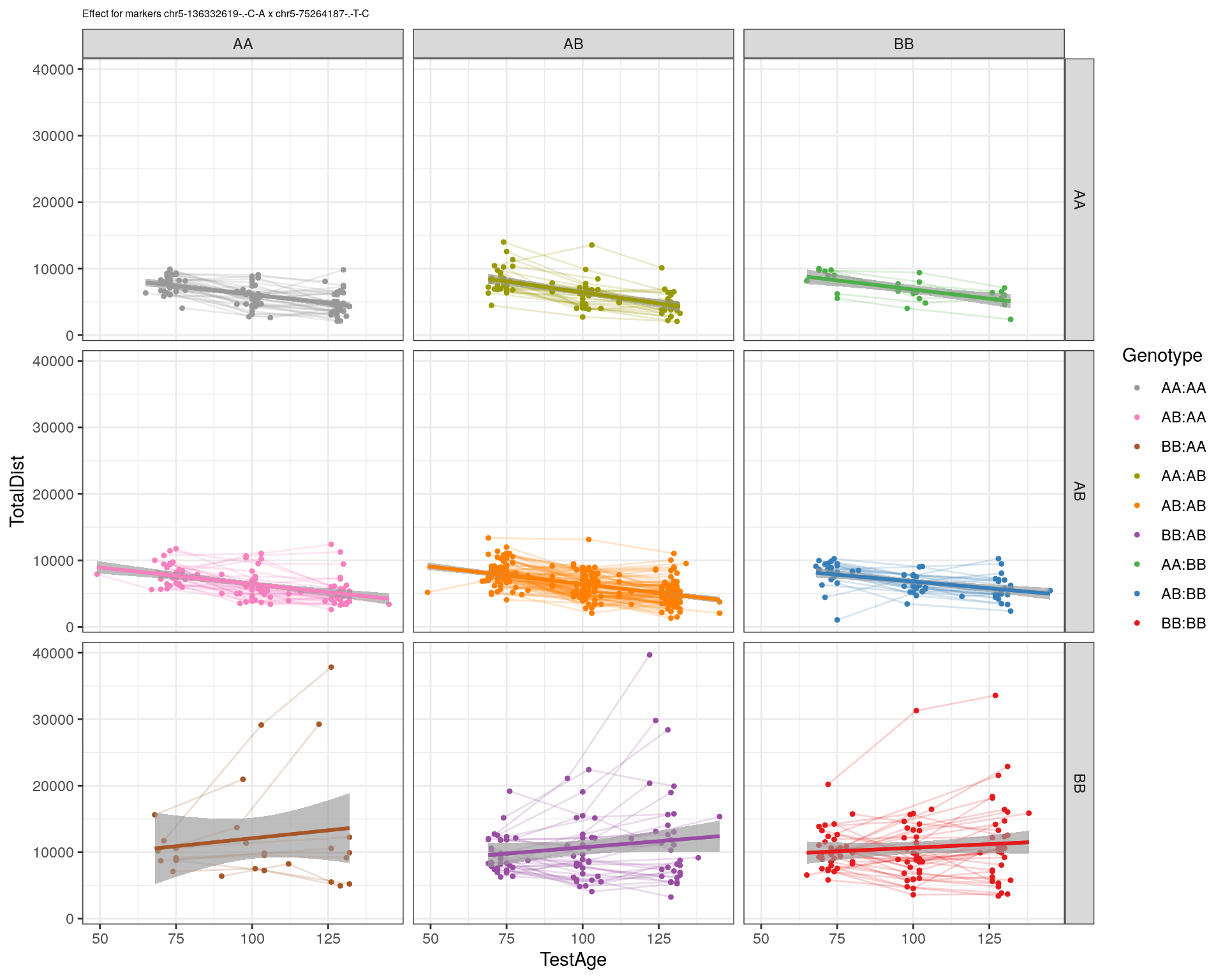
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-90233933-.-A-T"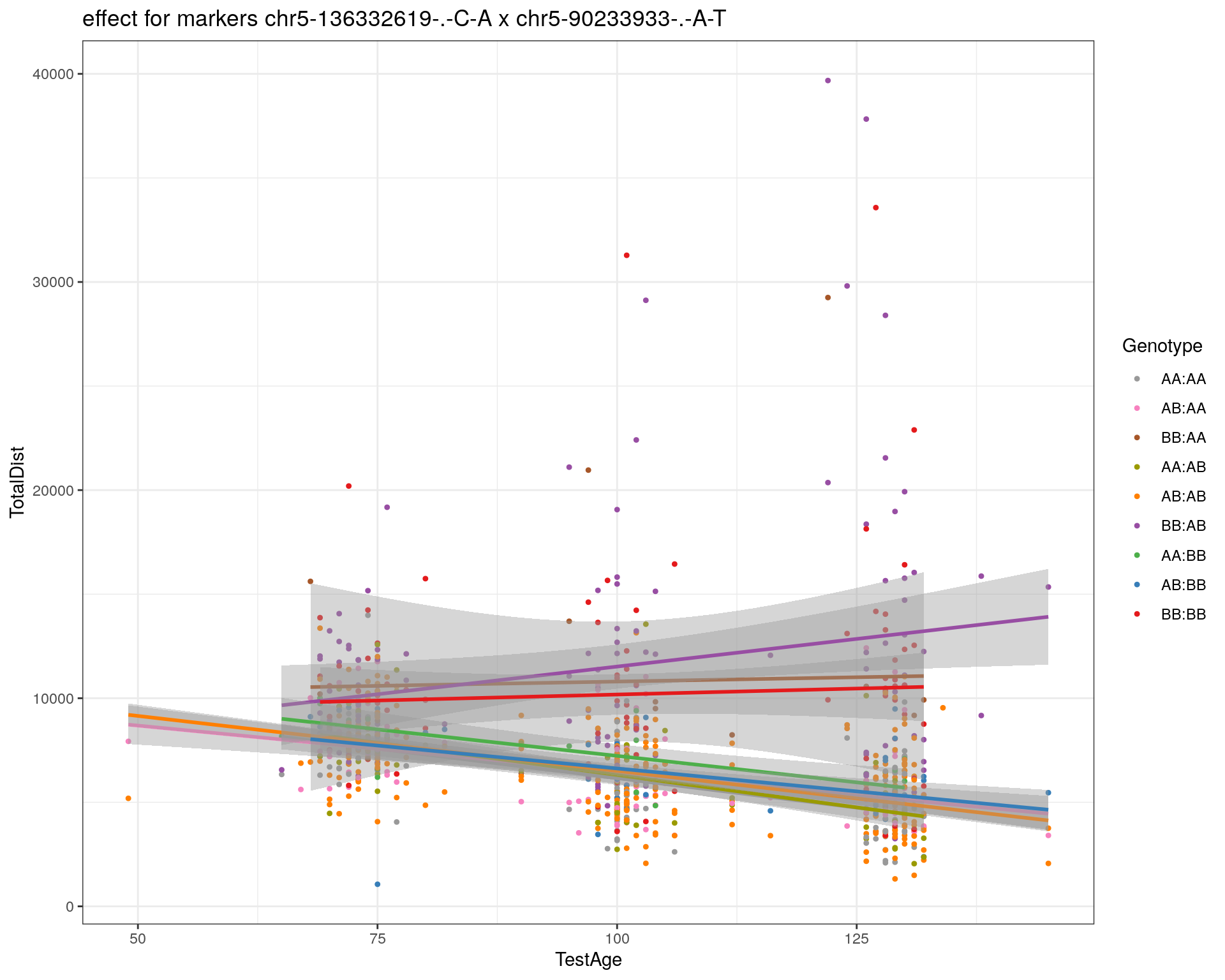
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
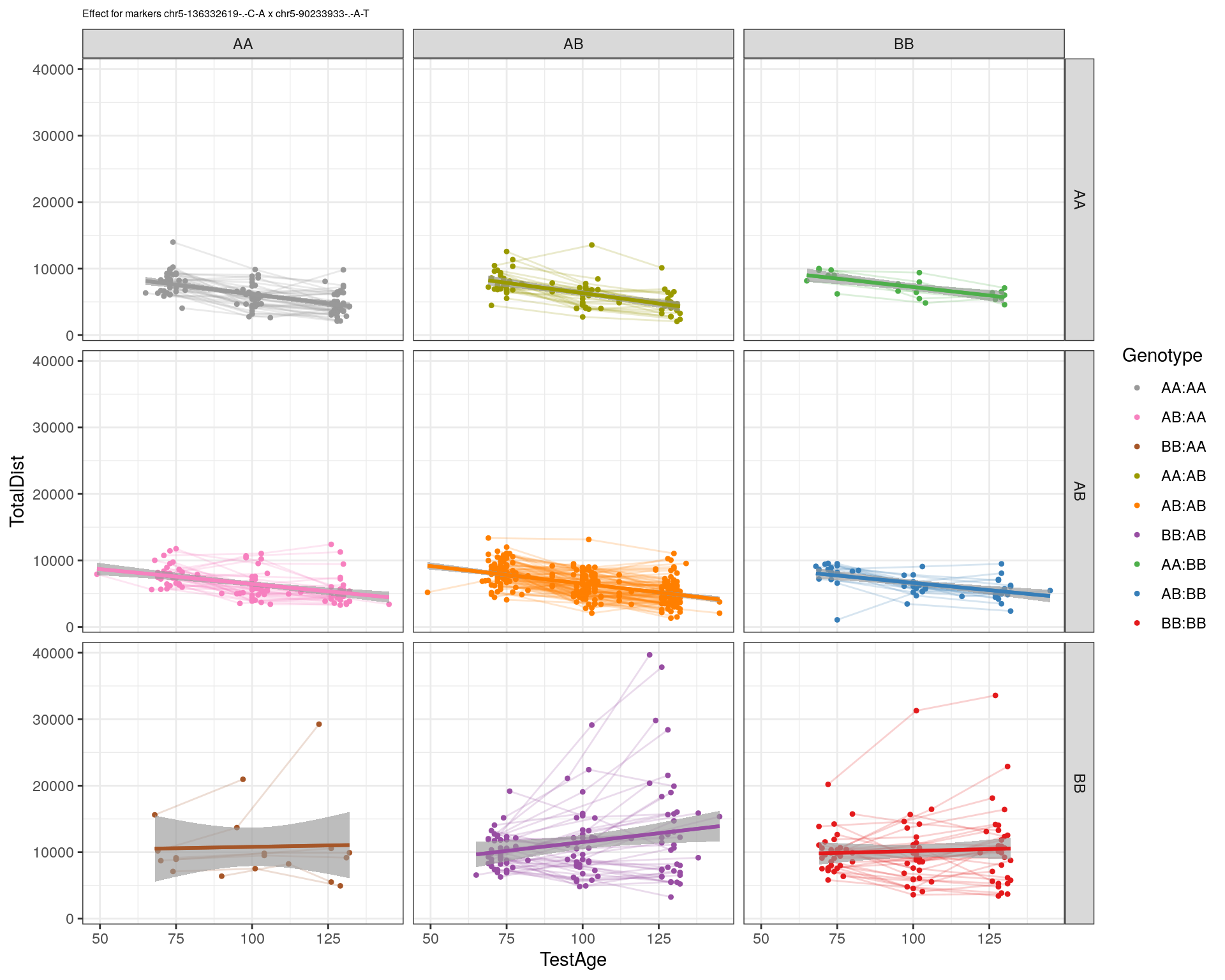
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-97607237-.-T-A"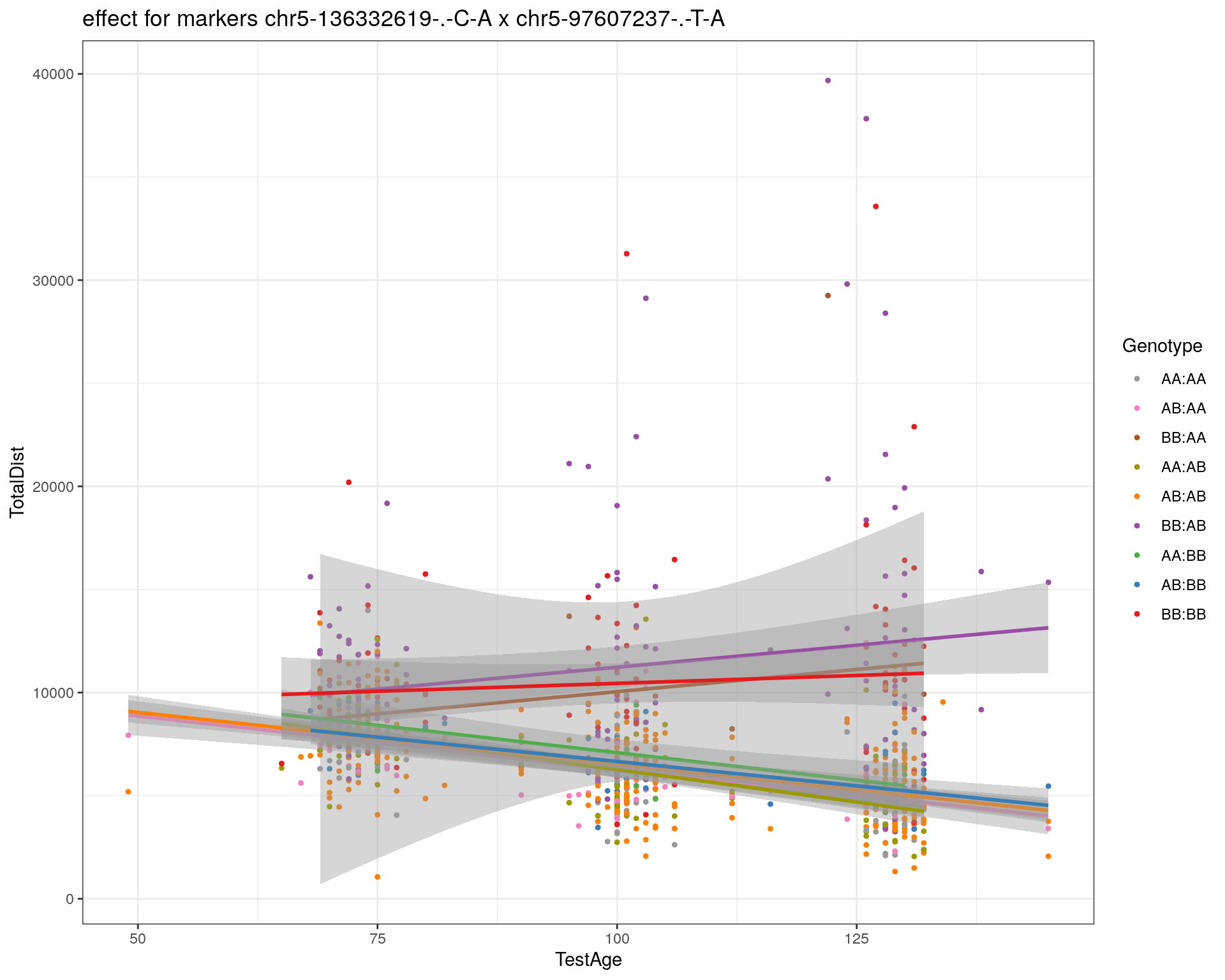
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
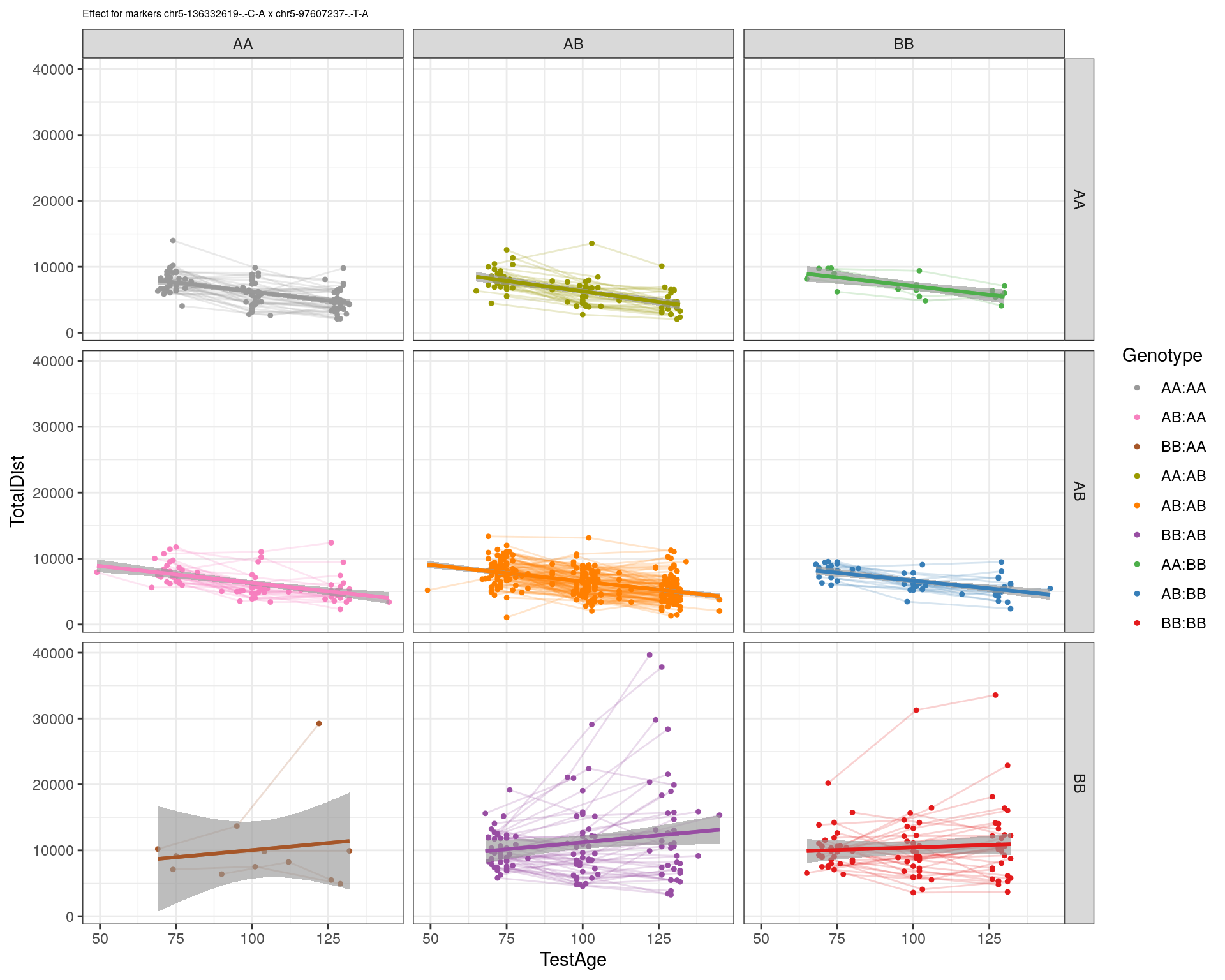
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-107507008-.-C-T"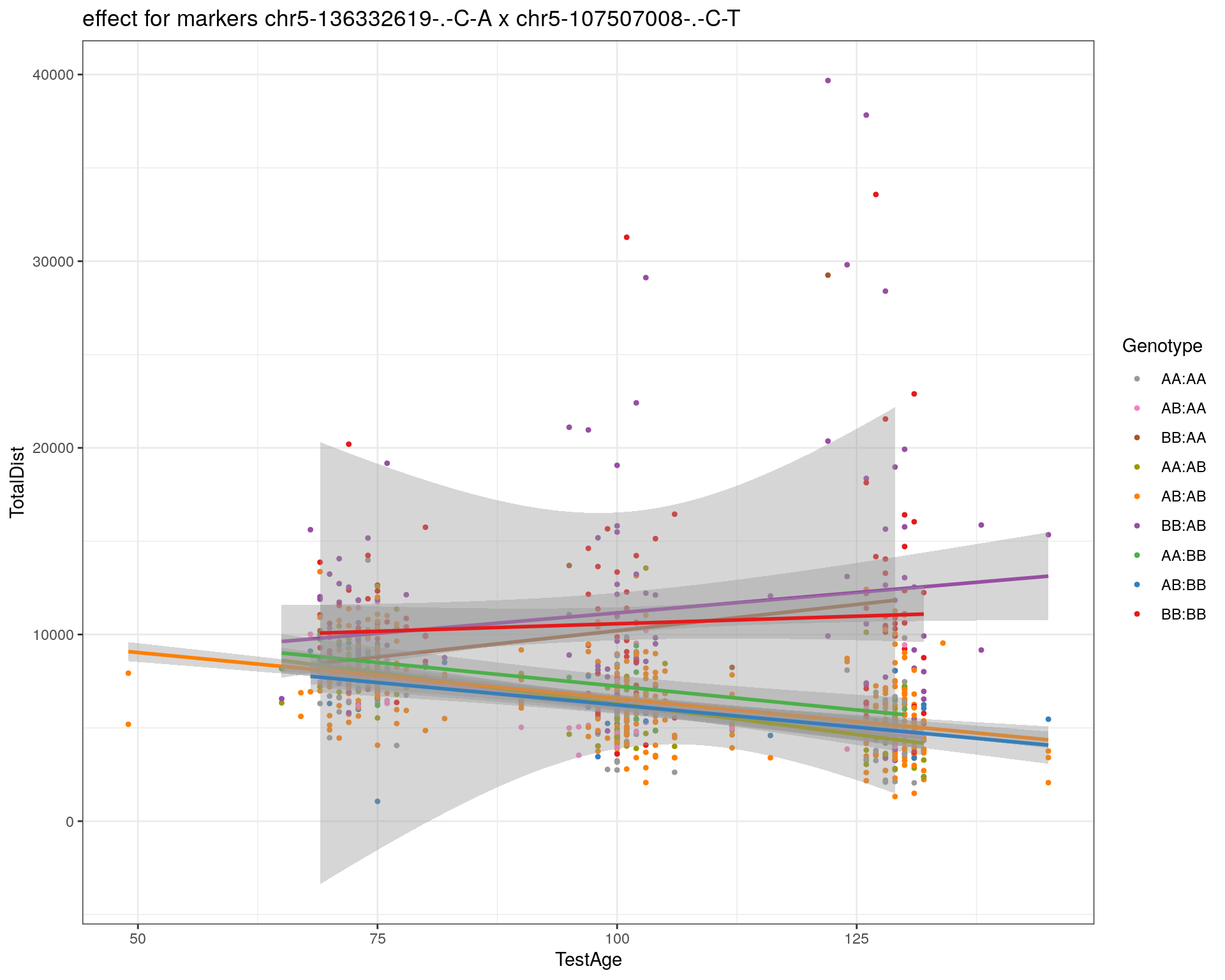
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
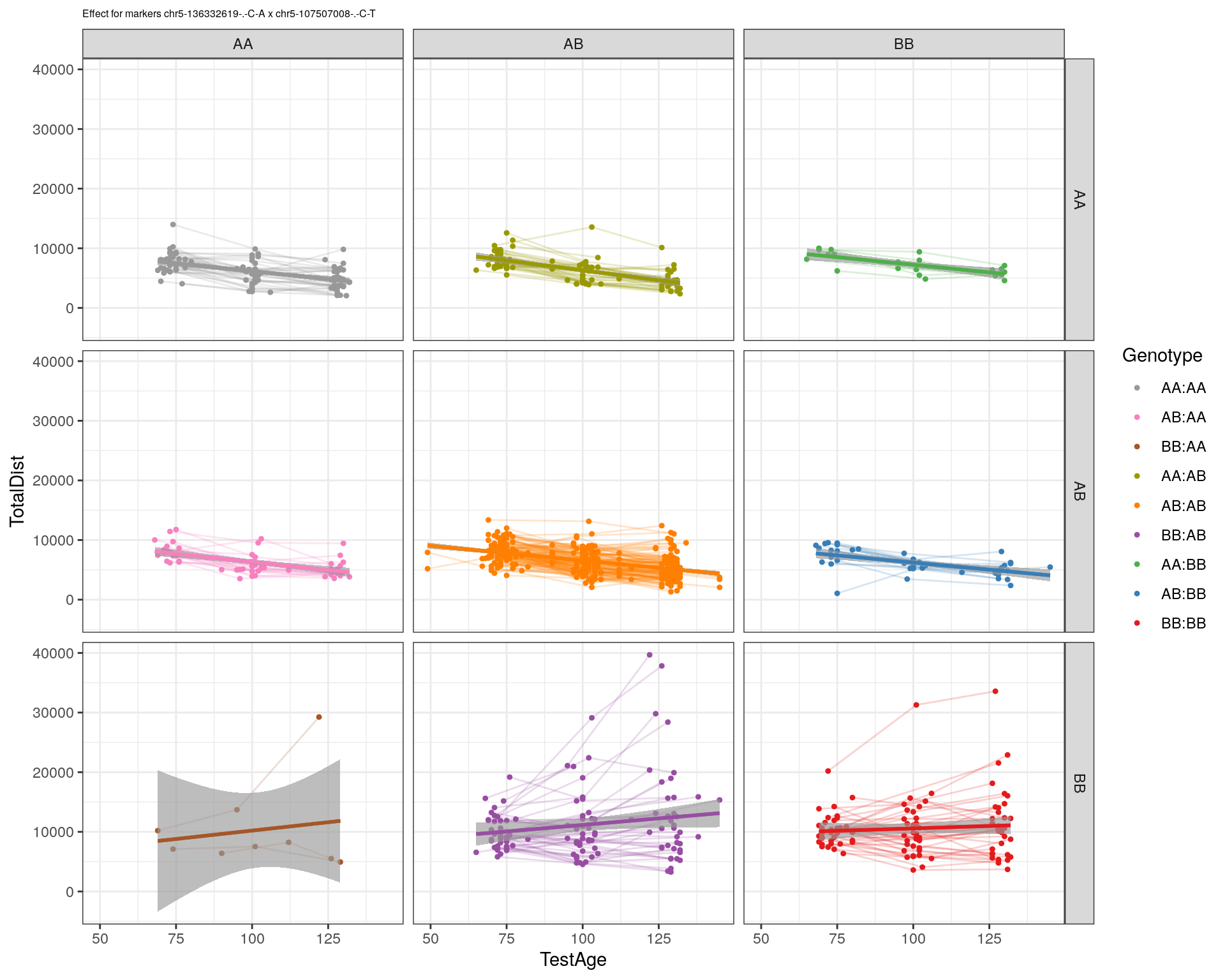
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-112799362-.-C-T"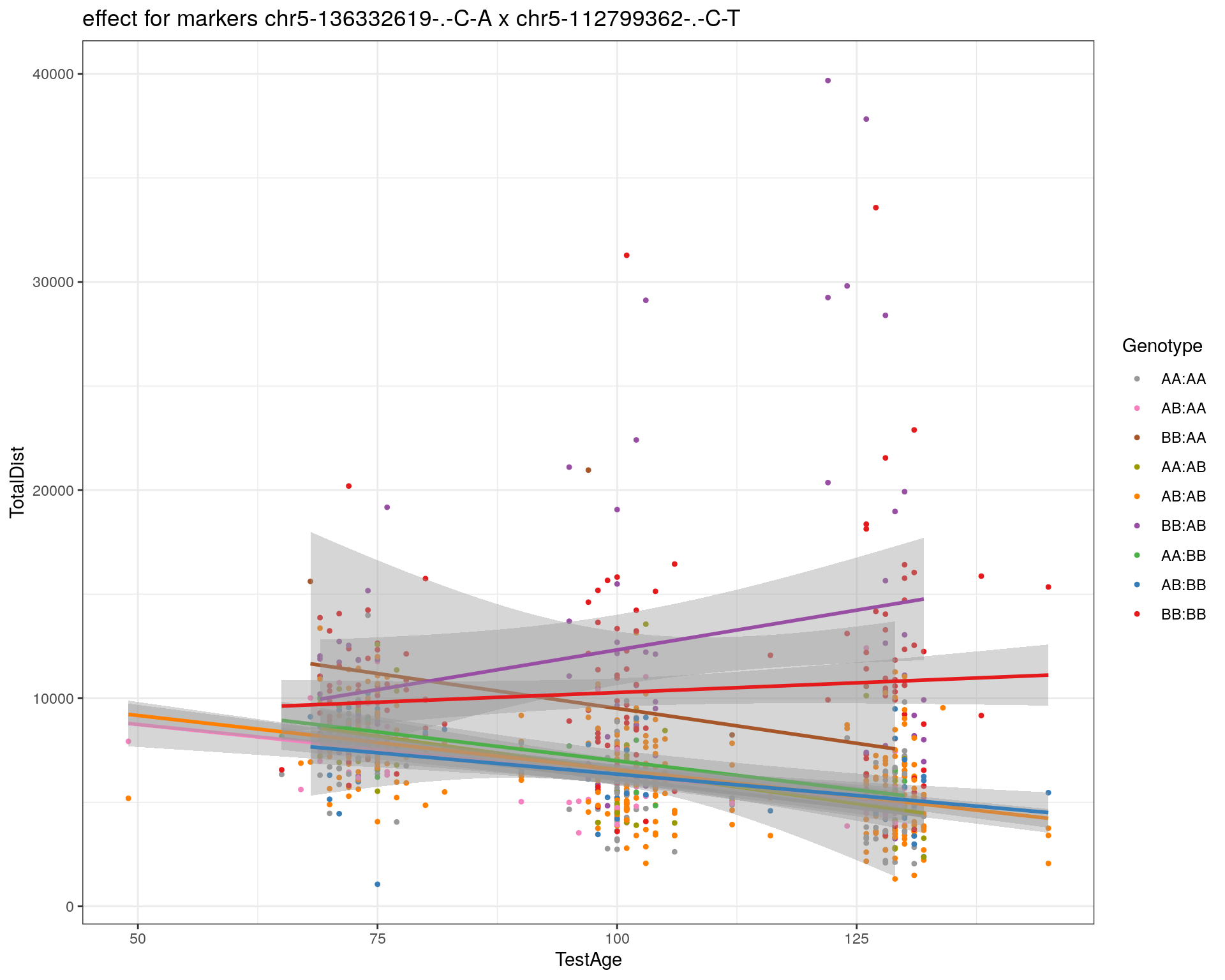
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
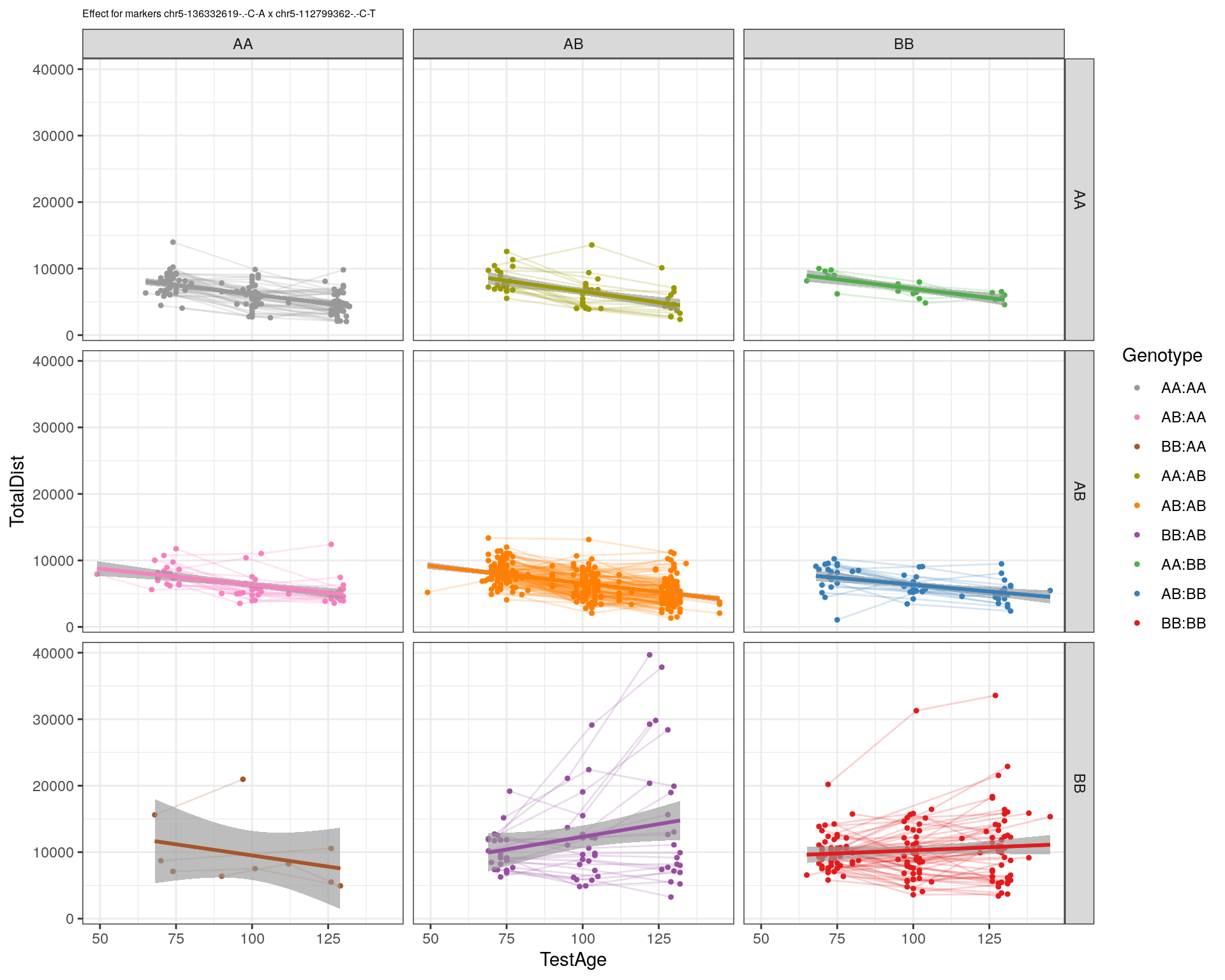
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-120660771-.-T-A"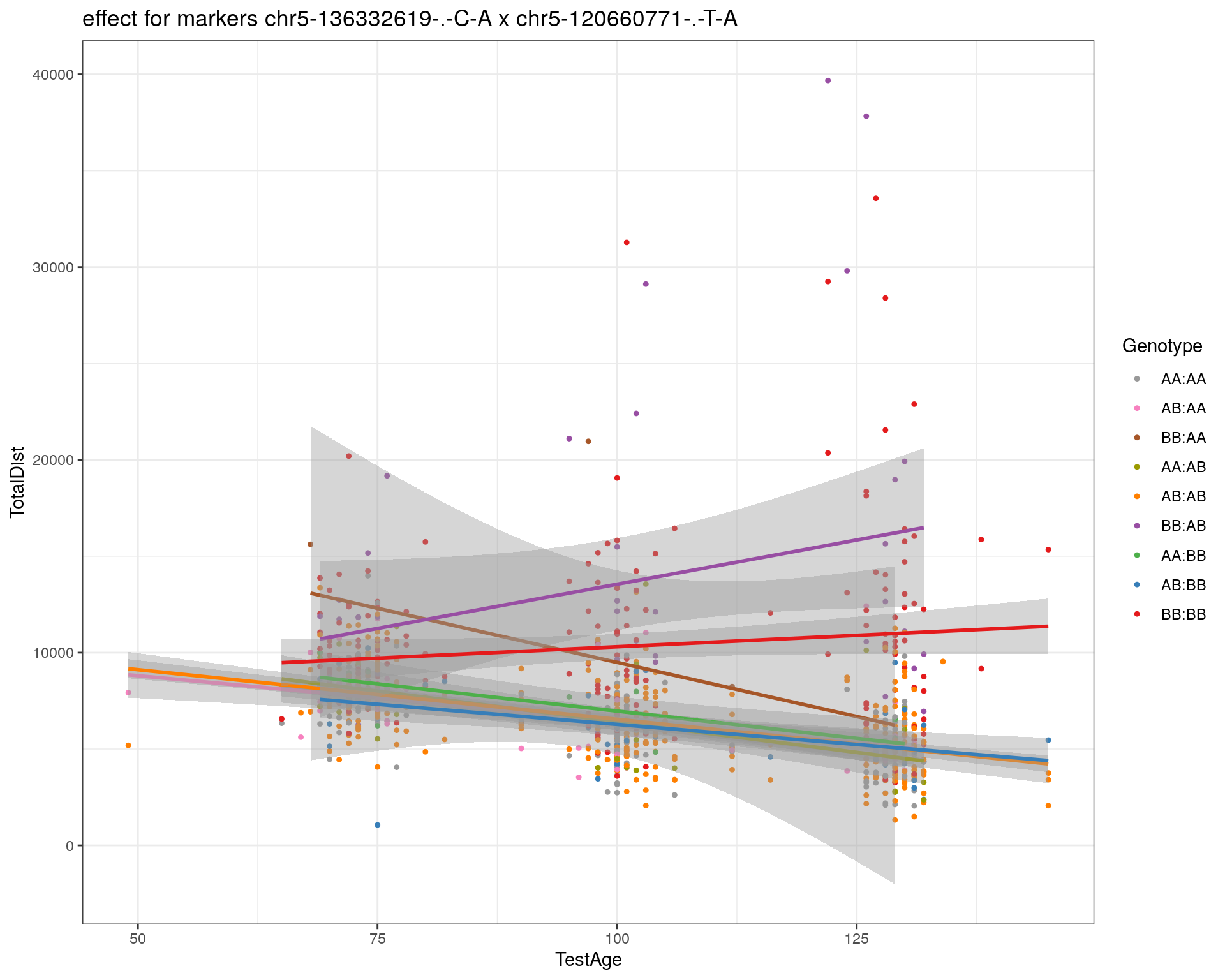
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
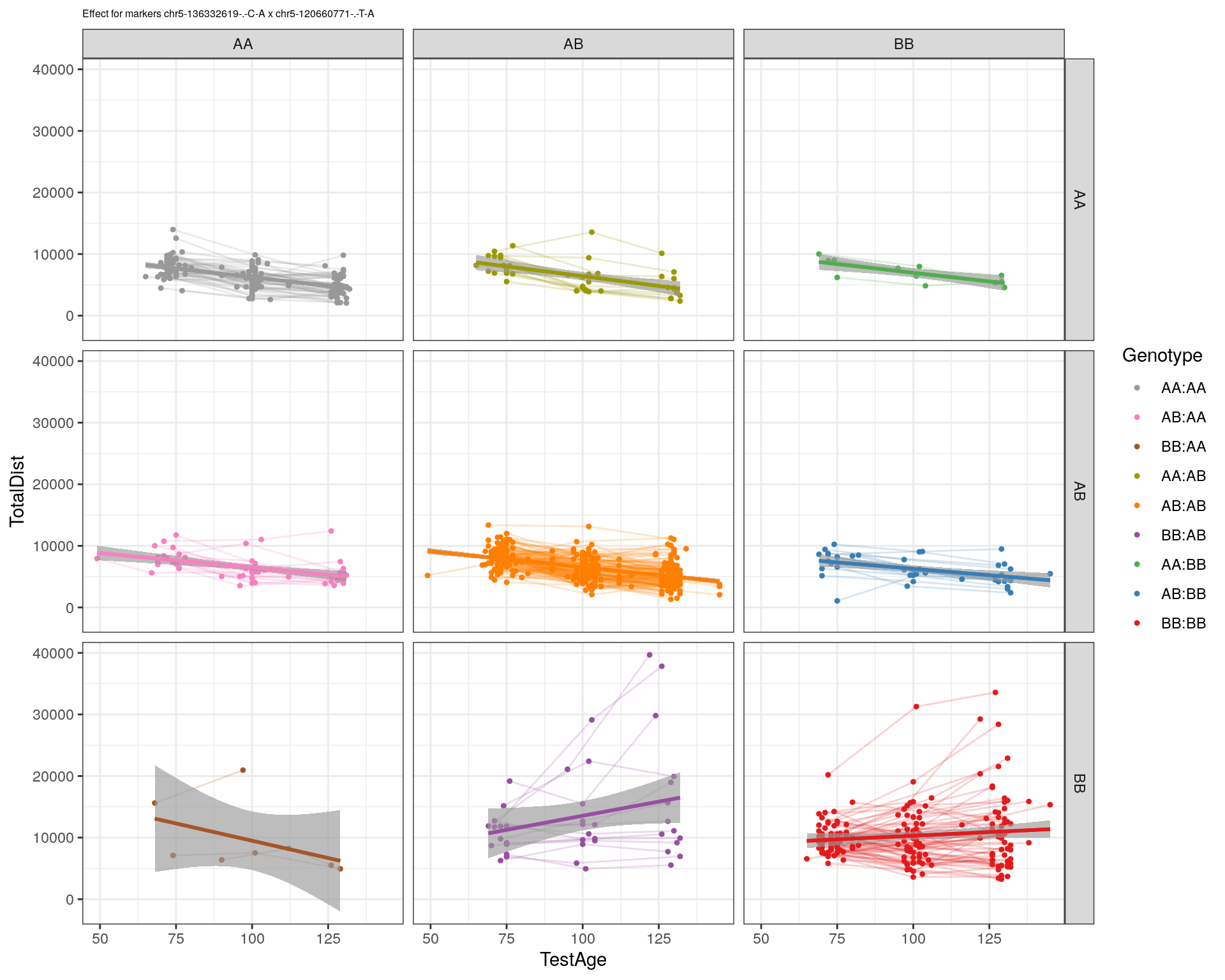
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-122167749-.-T-C"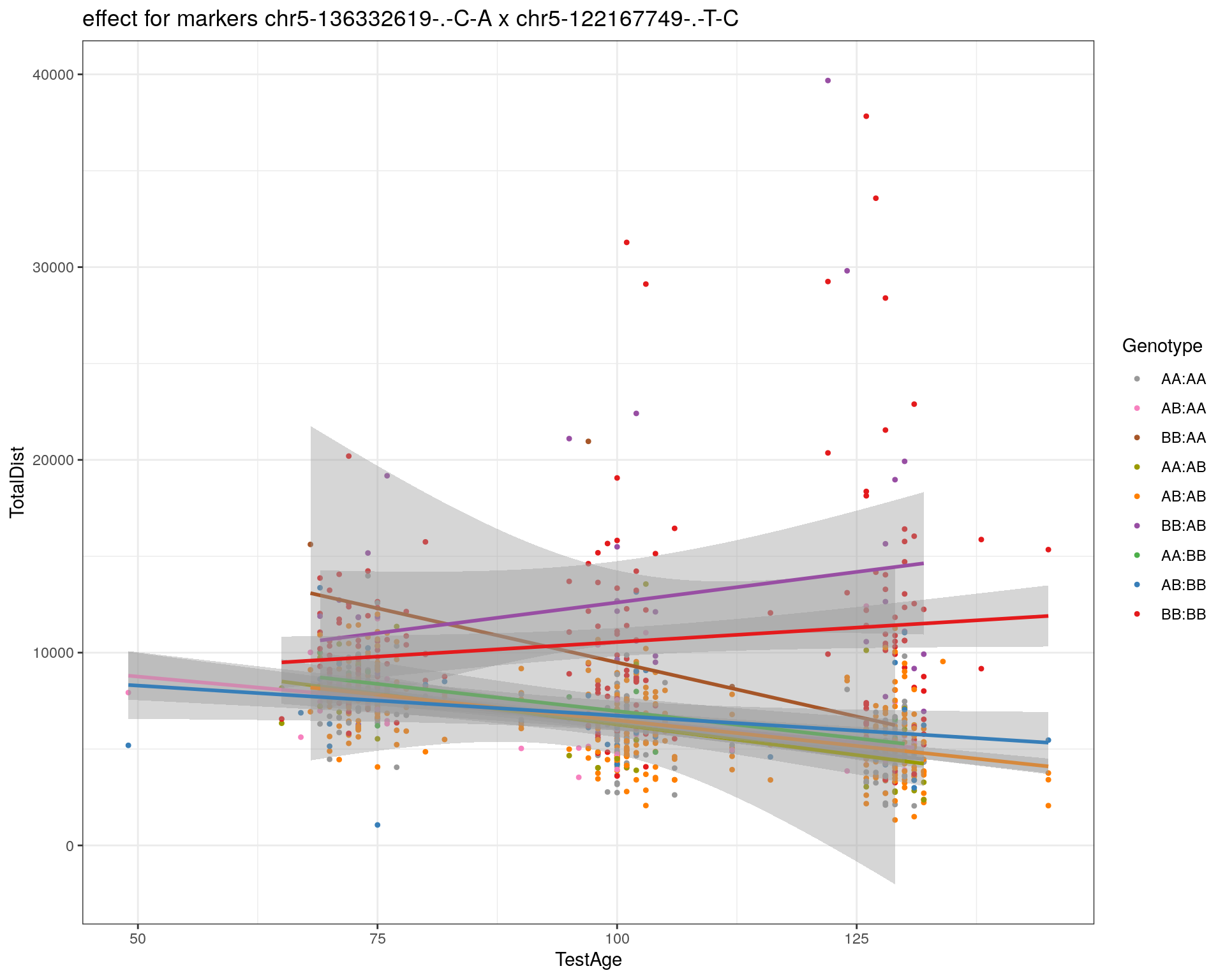
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
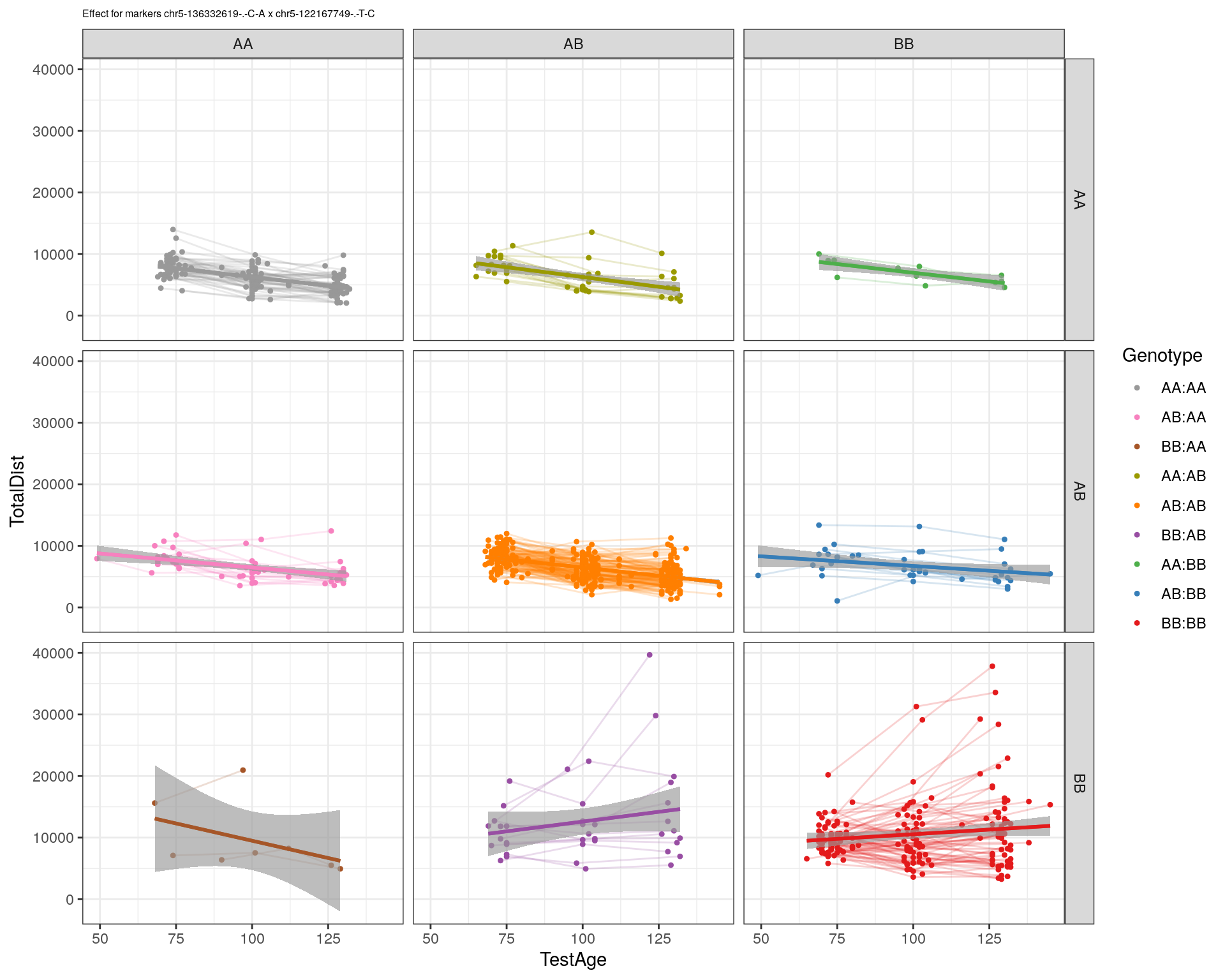
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-125153901-.-C-T"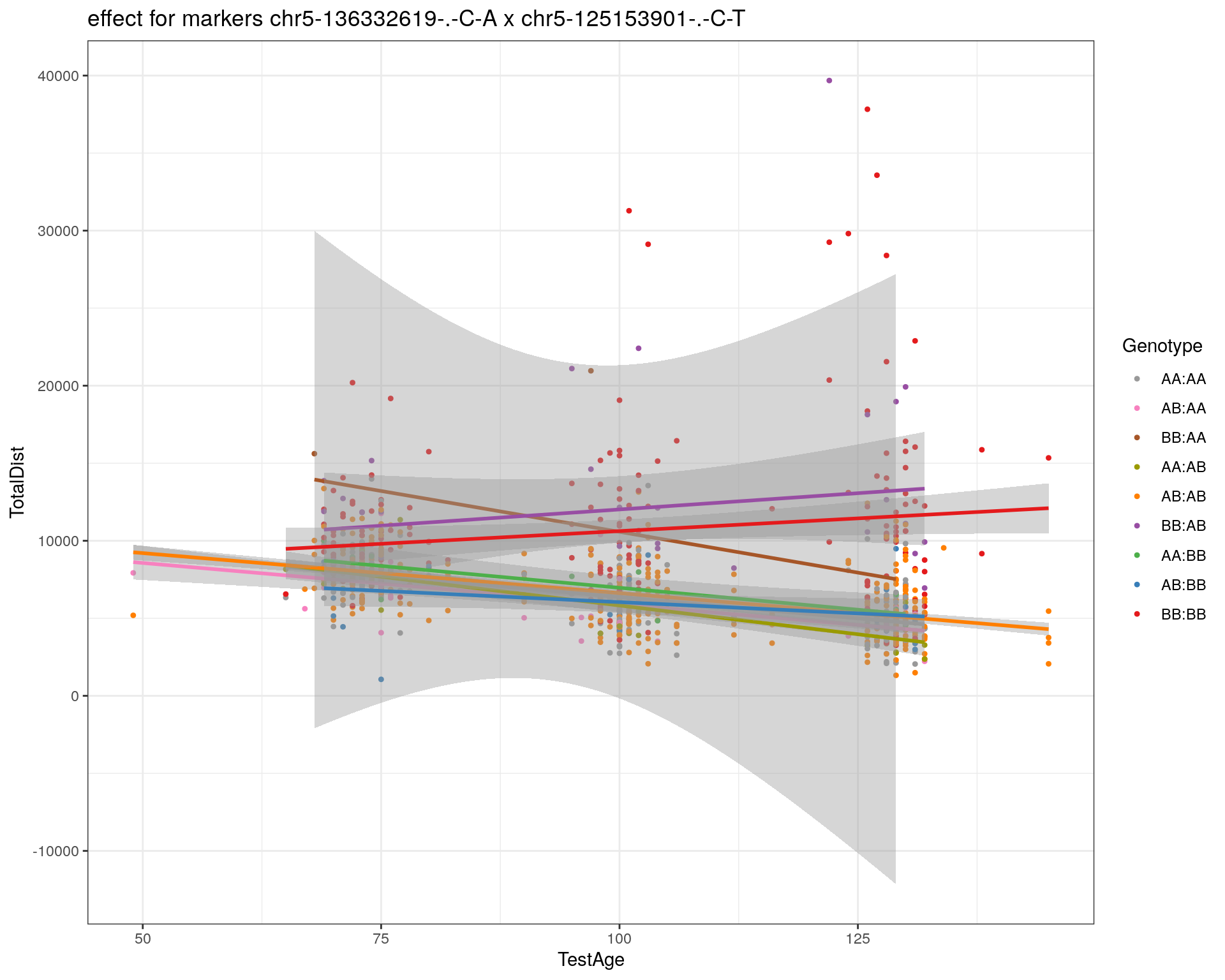
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-126058493-.-T-A"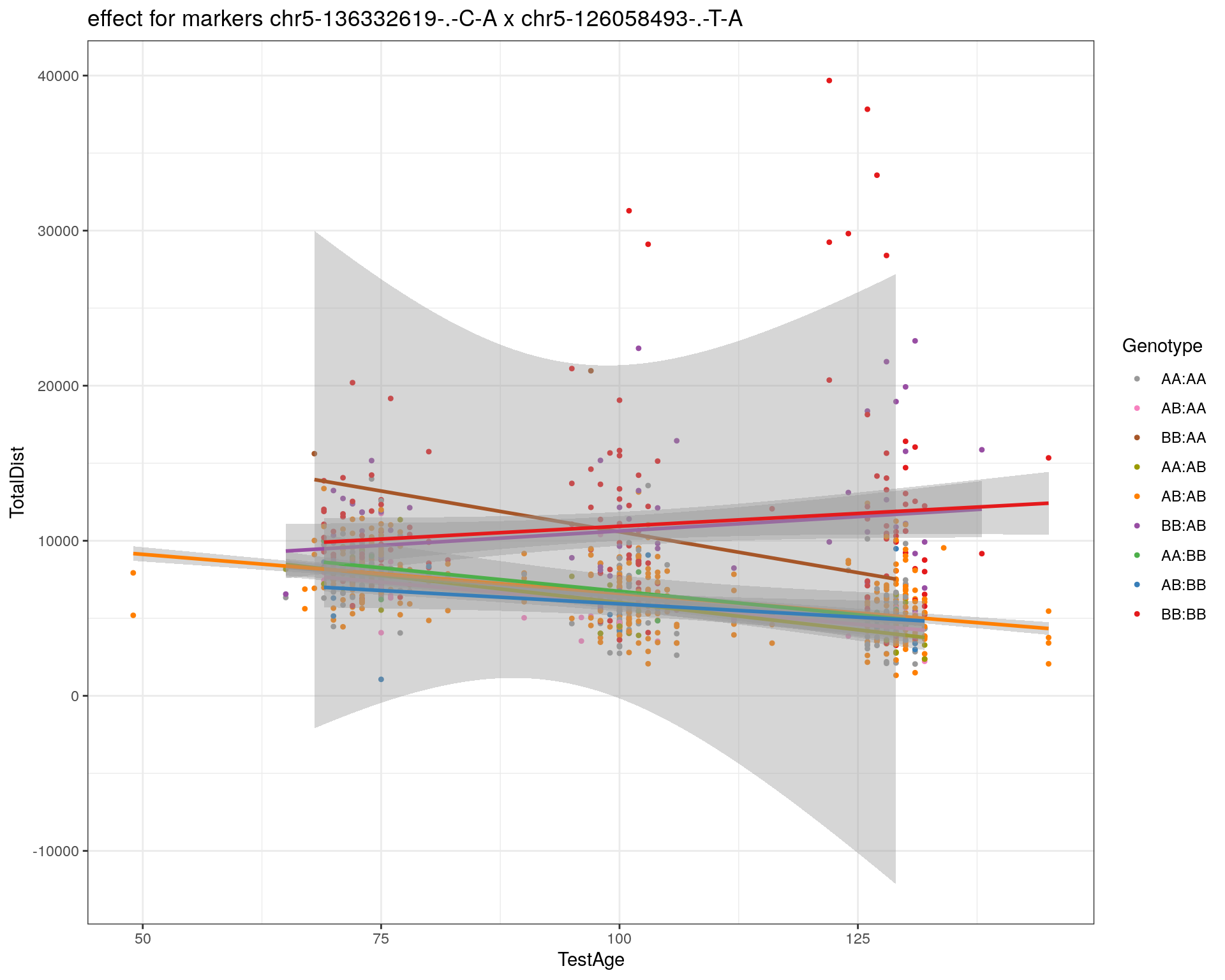
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-126551890-.-T-A"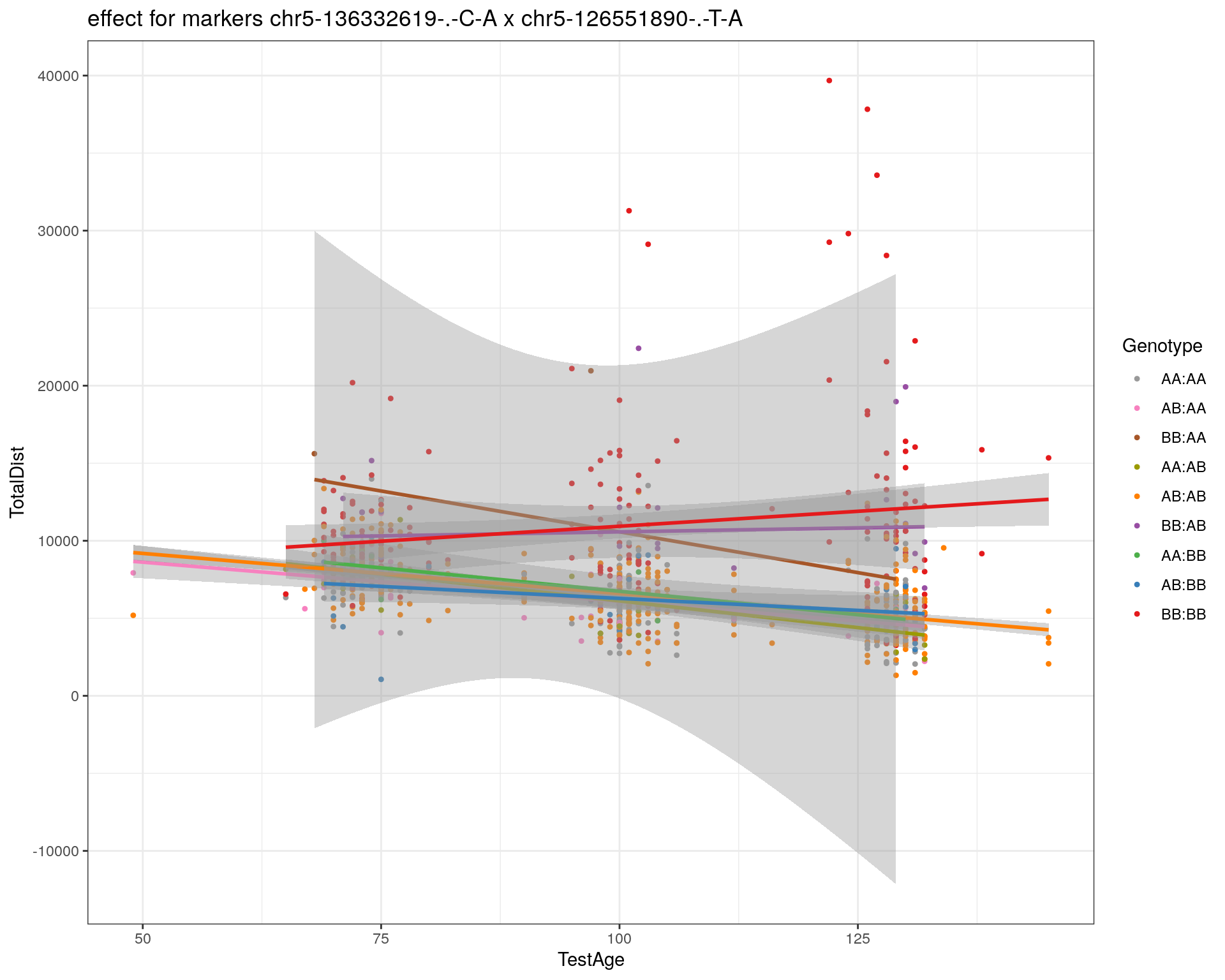
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
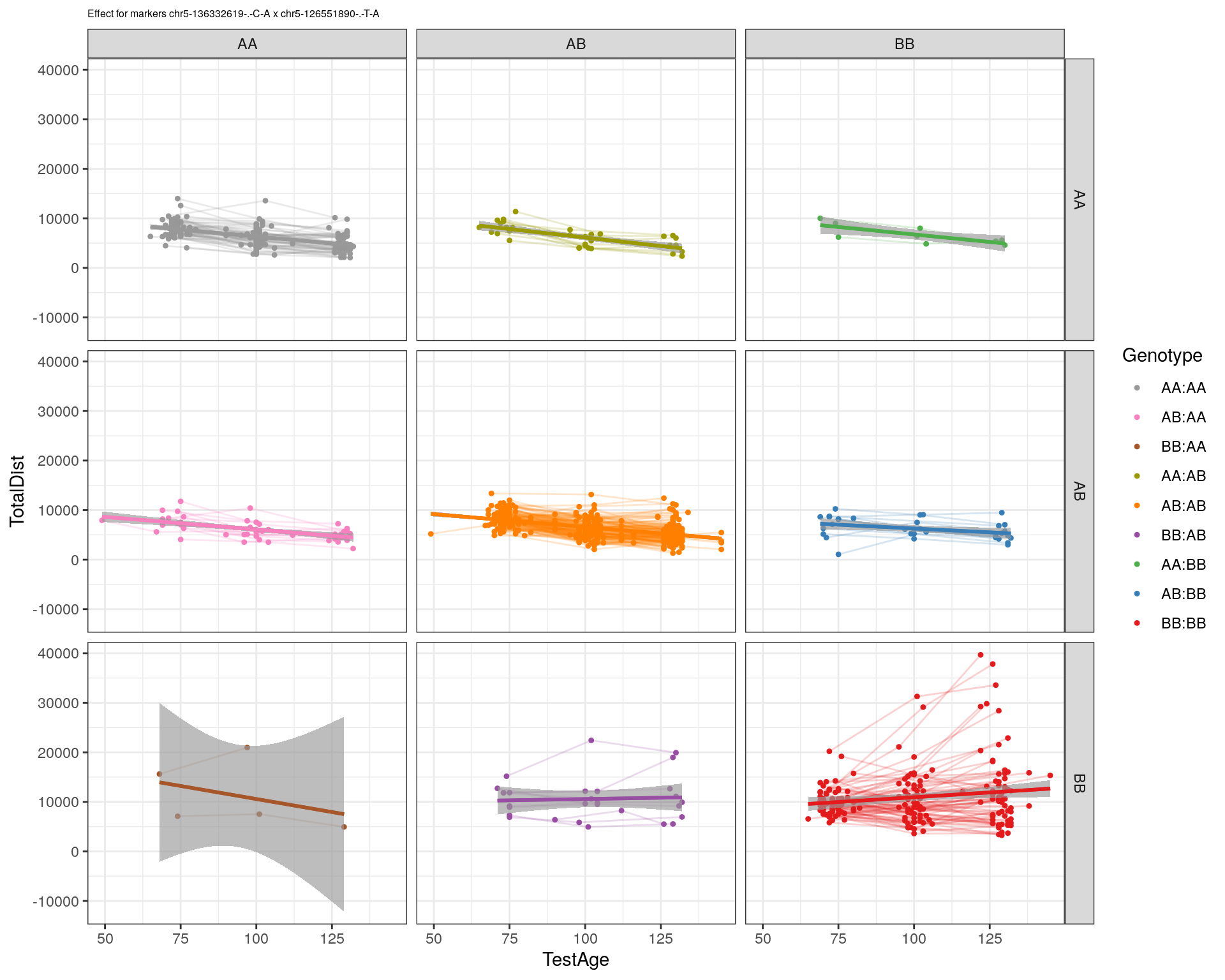
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-127565504-.-T-A"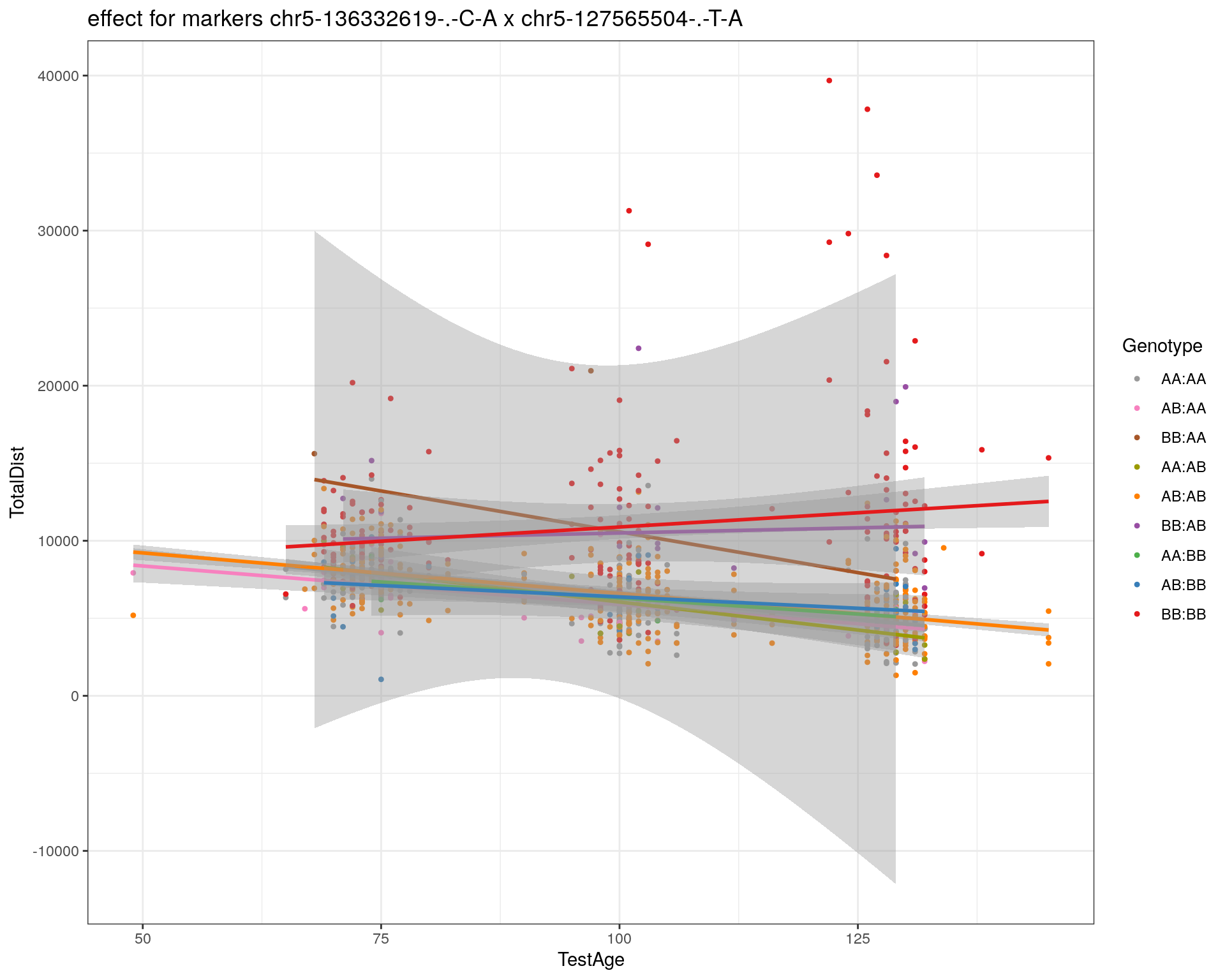
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
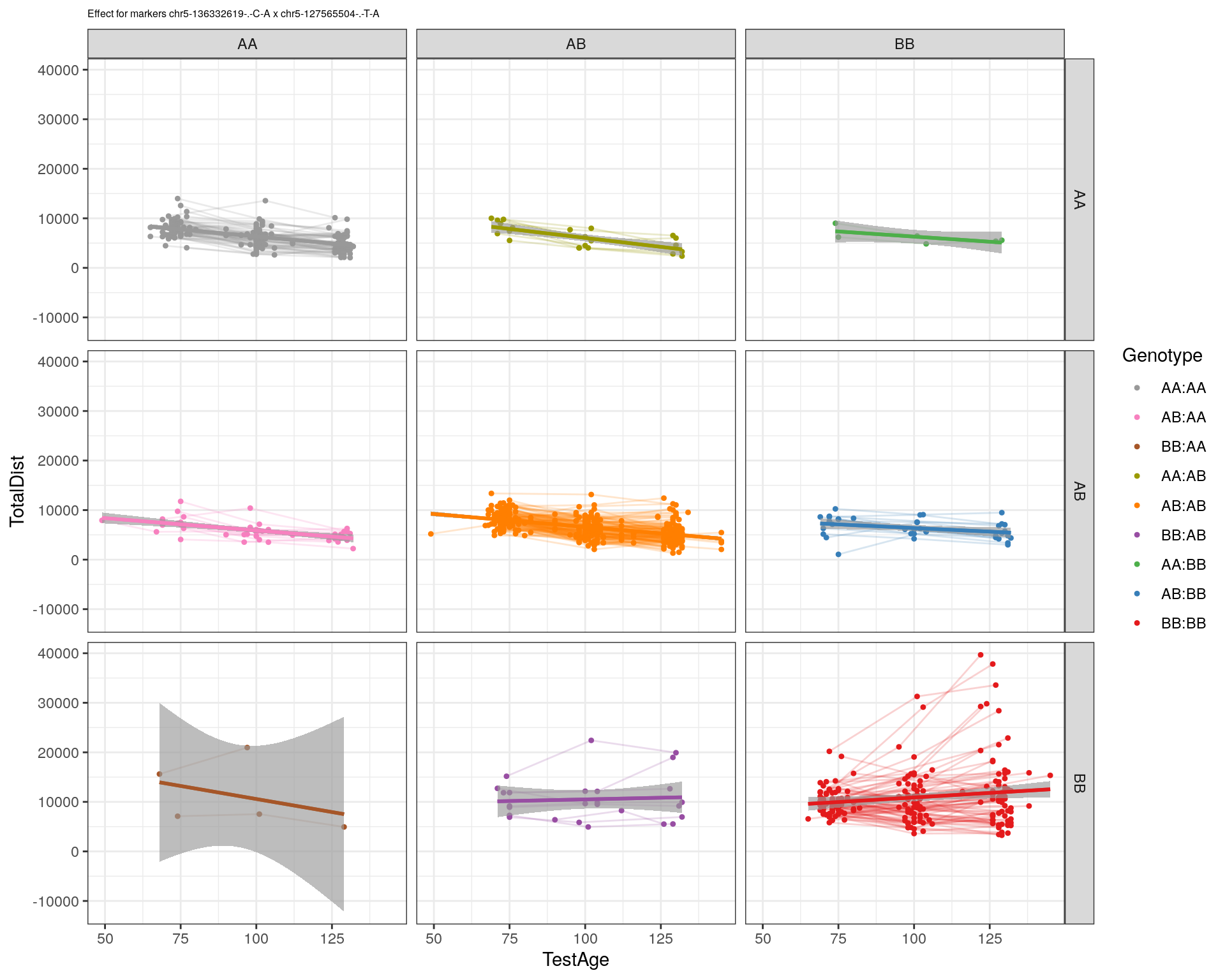
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-130394596-.-T-C"
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-130943677-.-T-A"
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-131298836-.-T-C"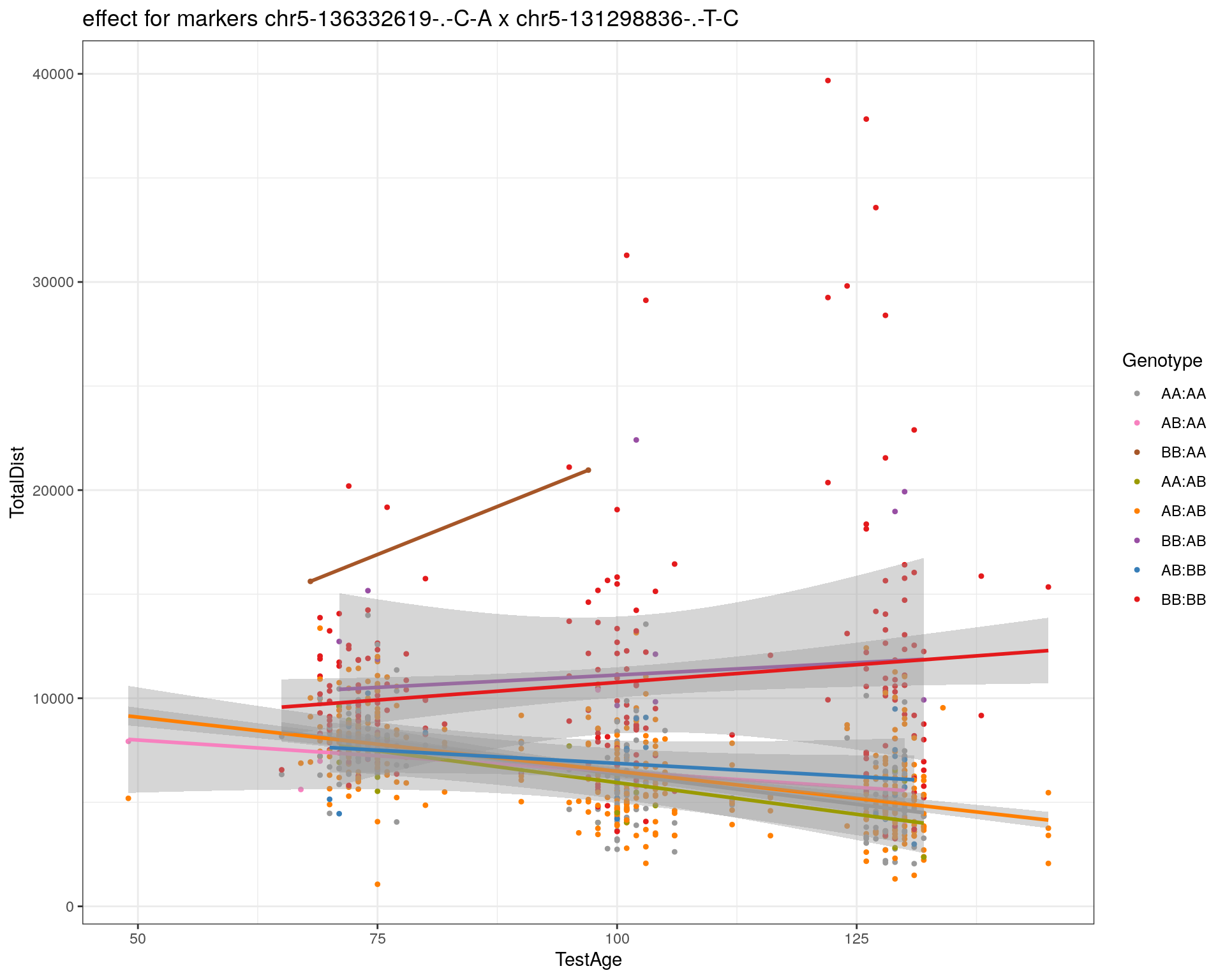
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
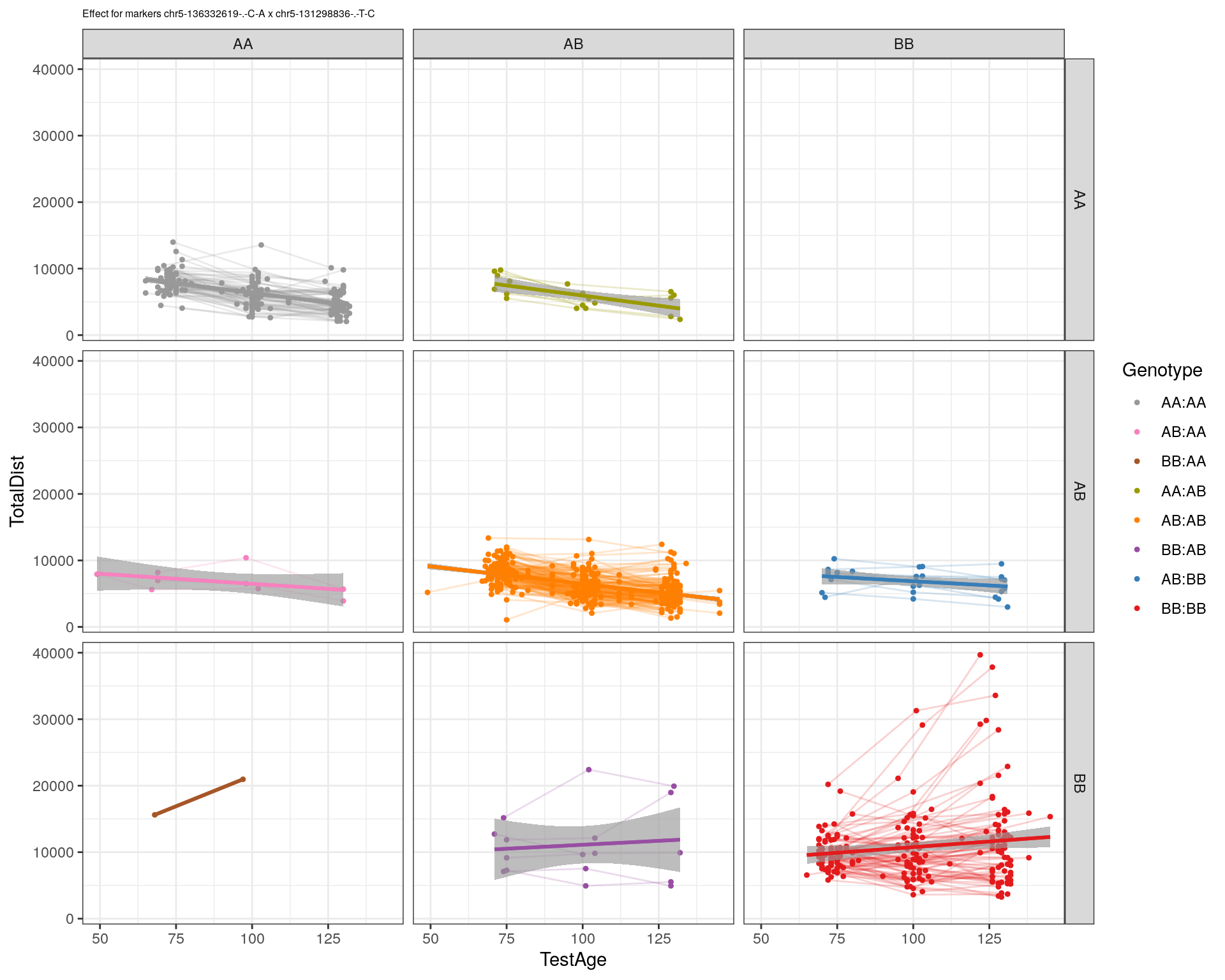
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-133052642-.-T-G"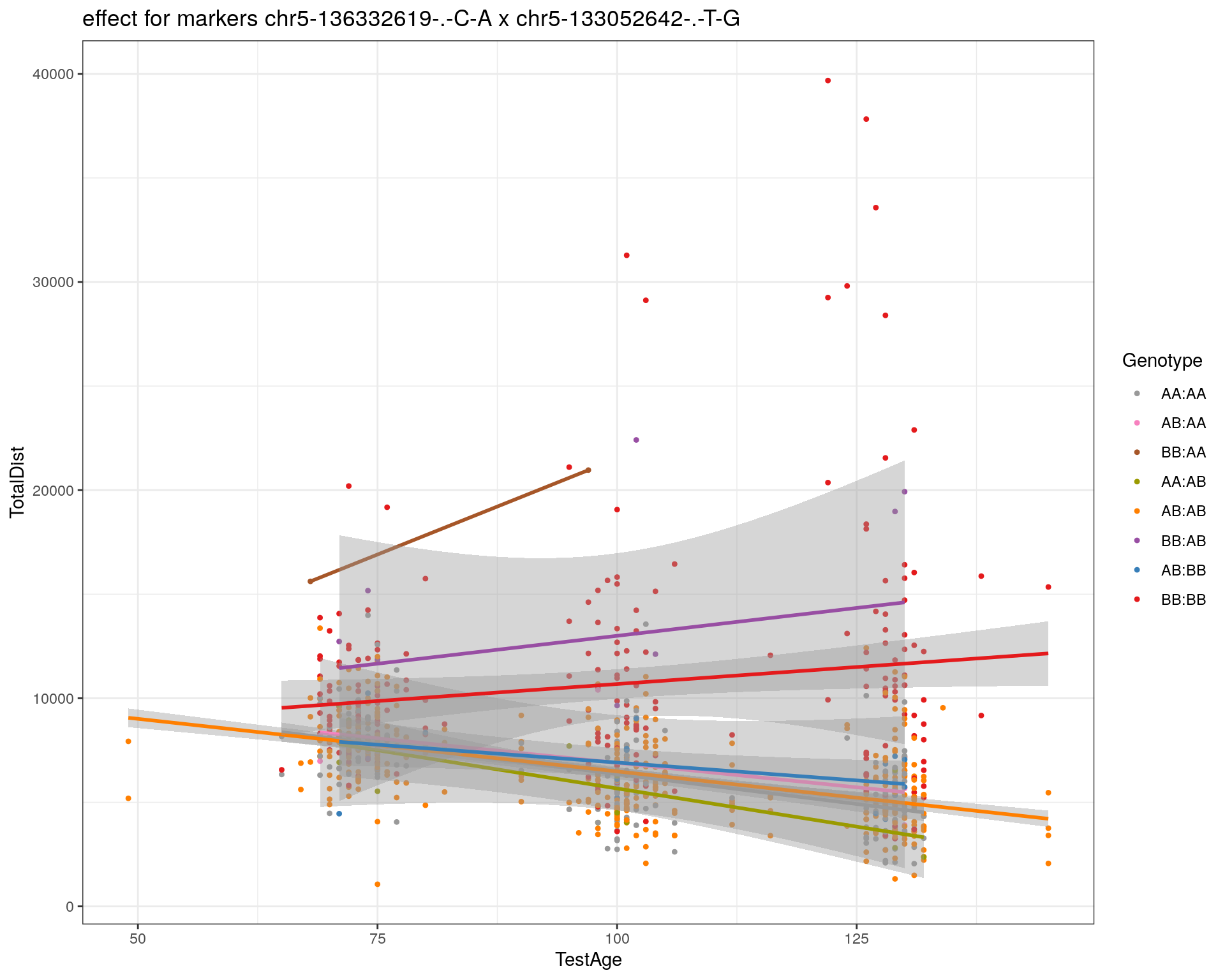
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
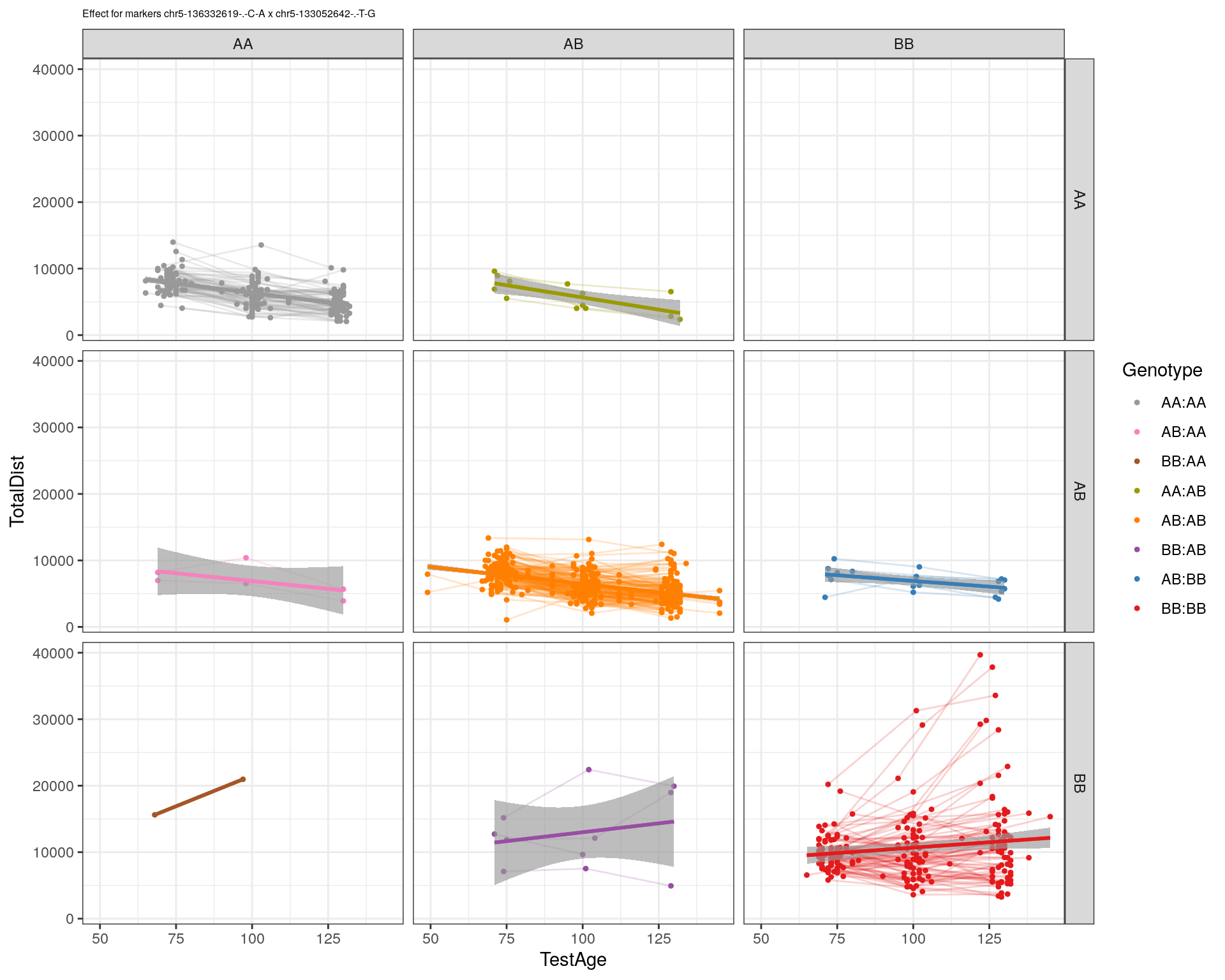
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
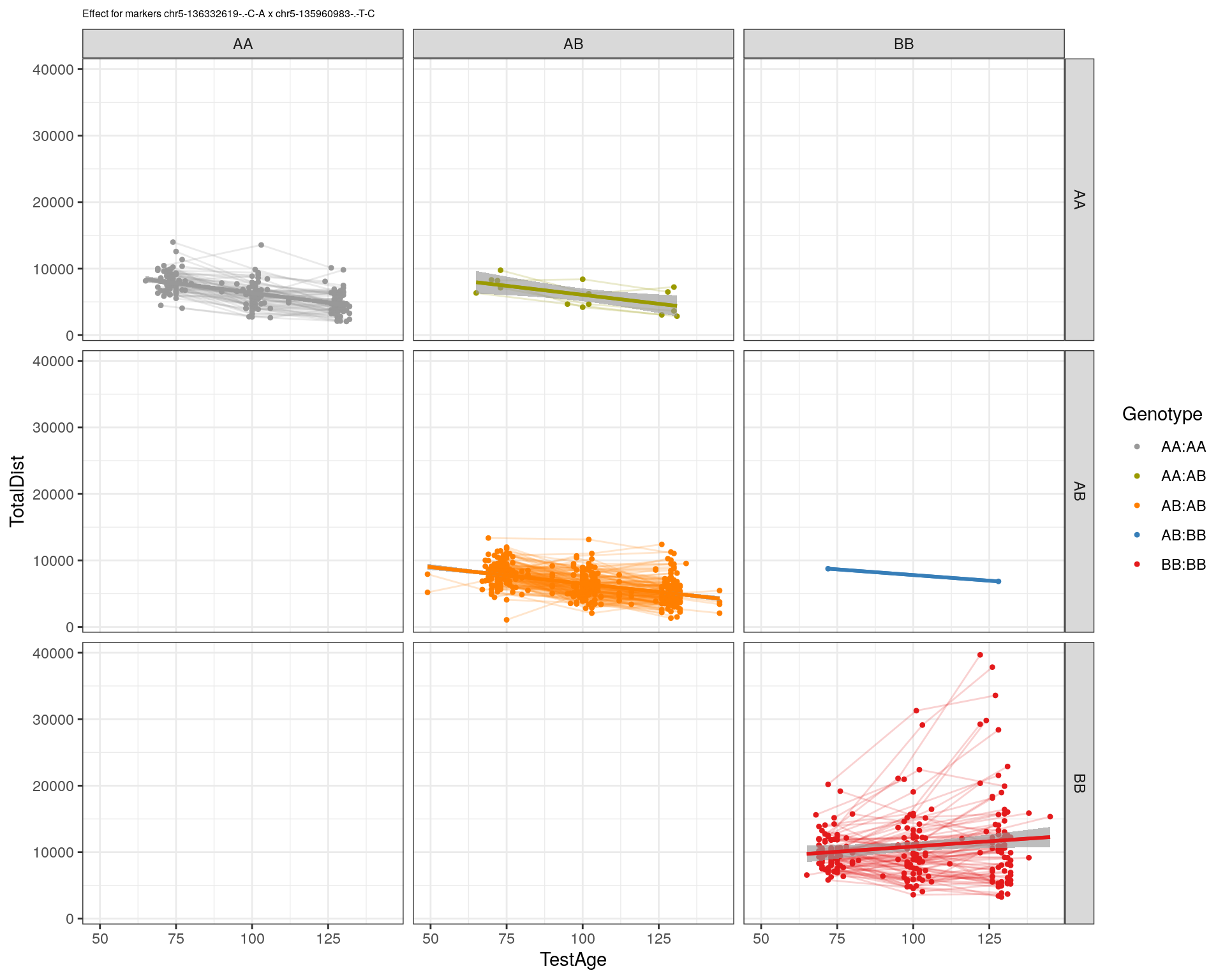
[1] "chr5-135960983-.-T-C"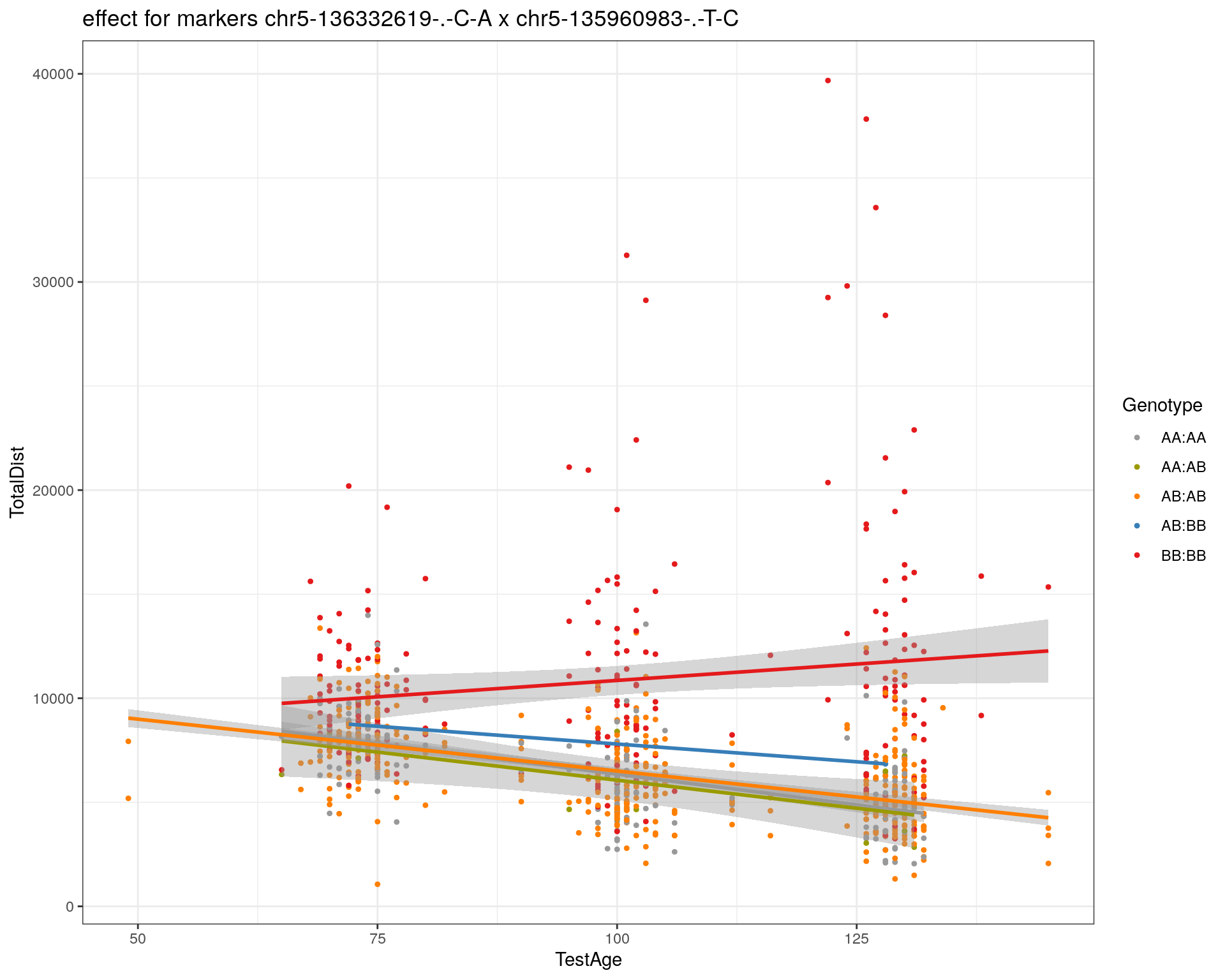
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
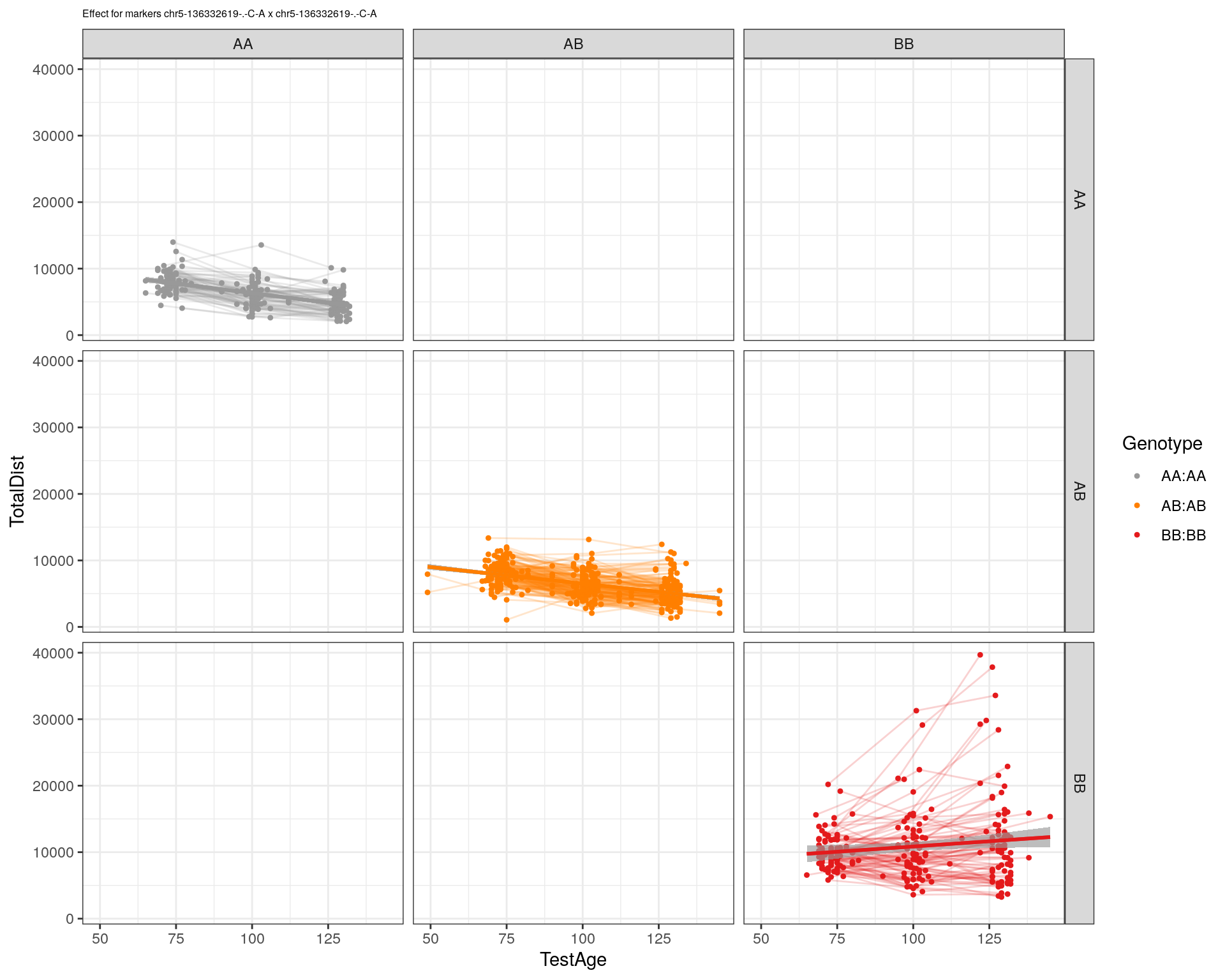
[1] "chr5-136332619-.-C-A"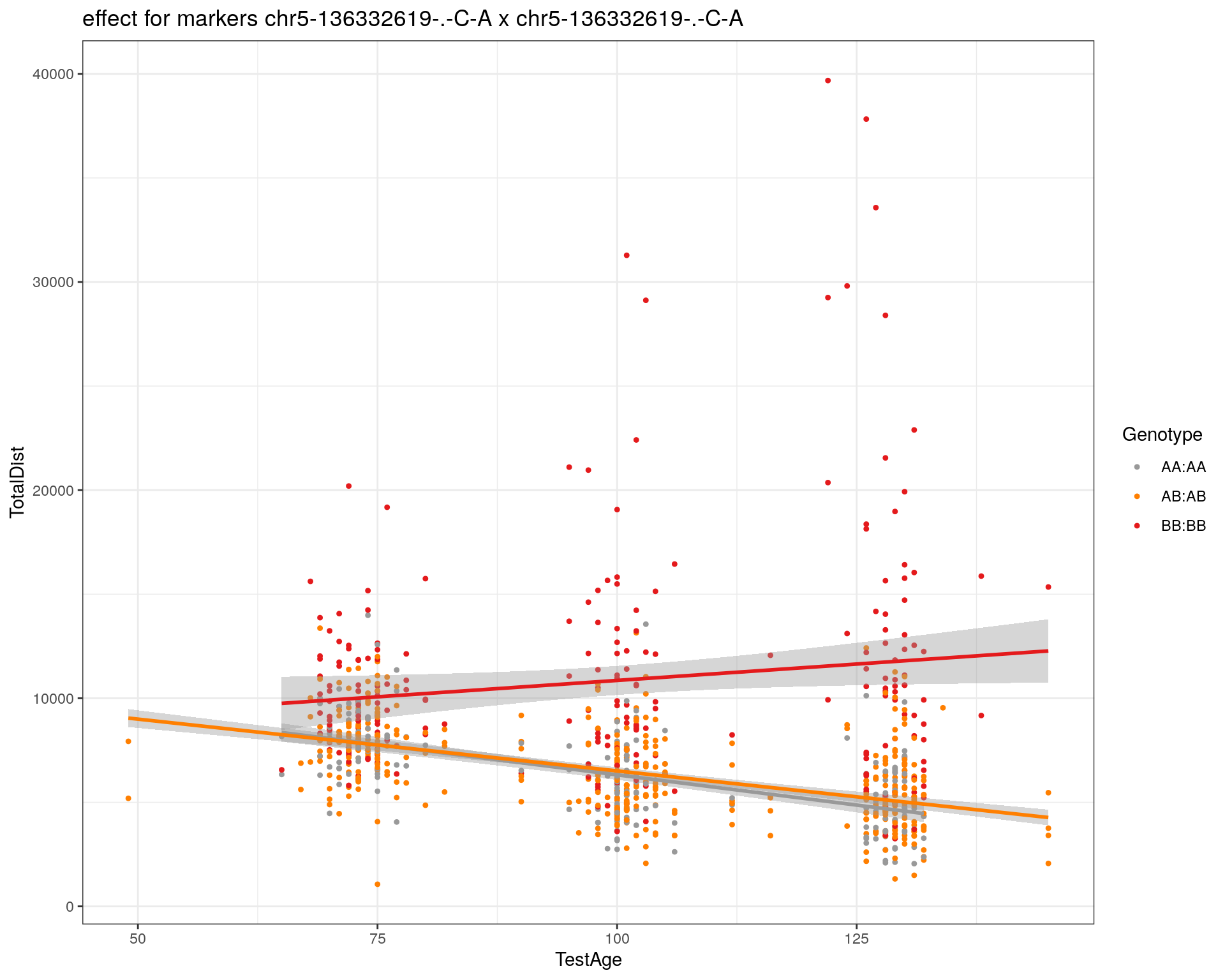
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
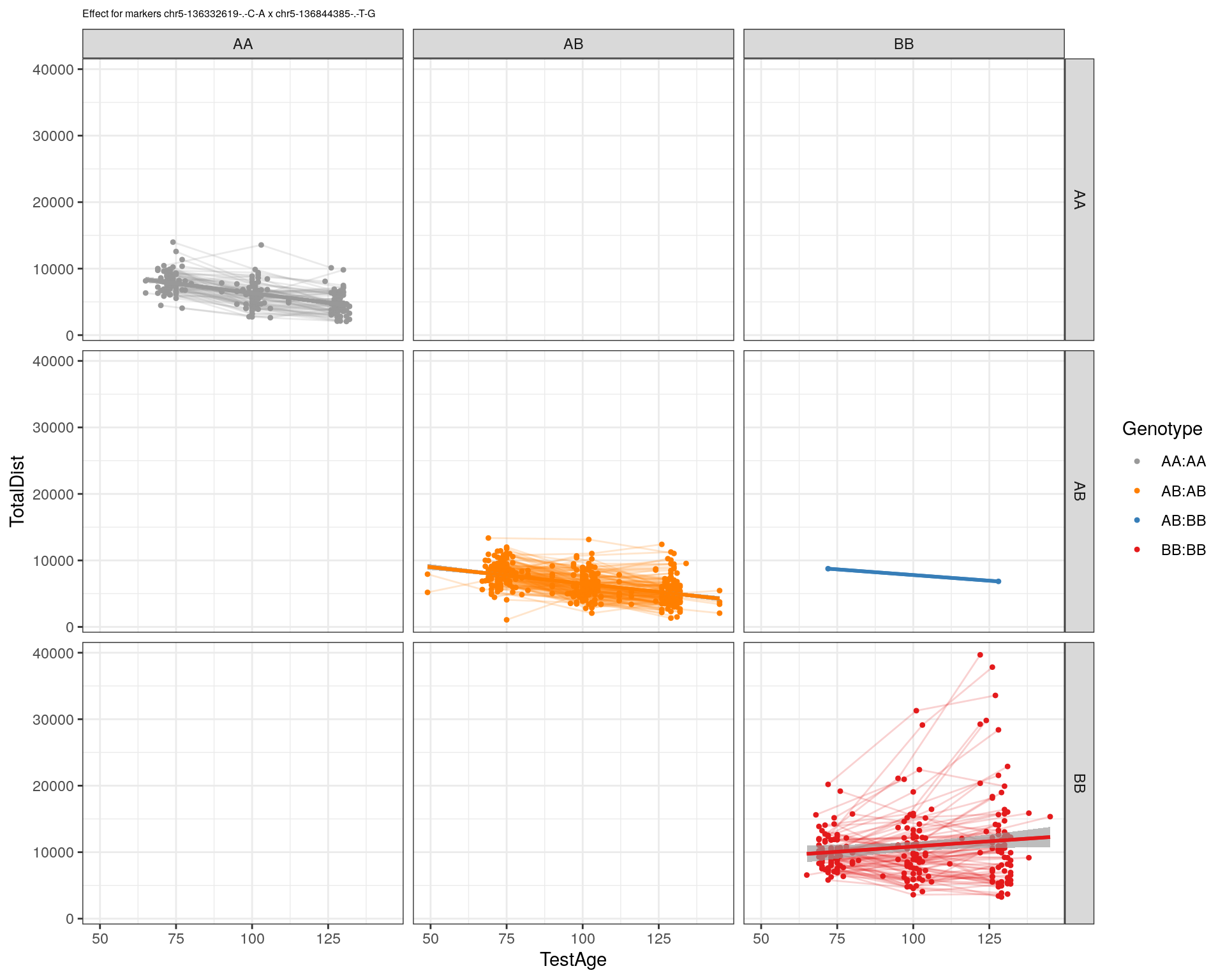
[1] "chr5-136844385-.-T-G"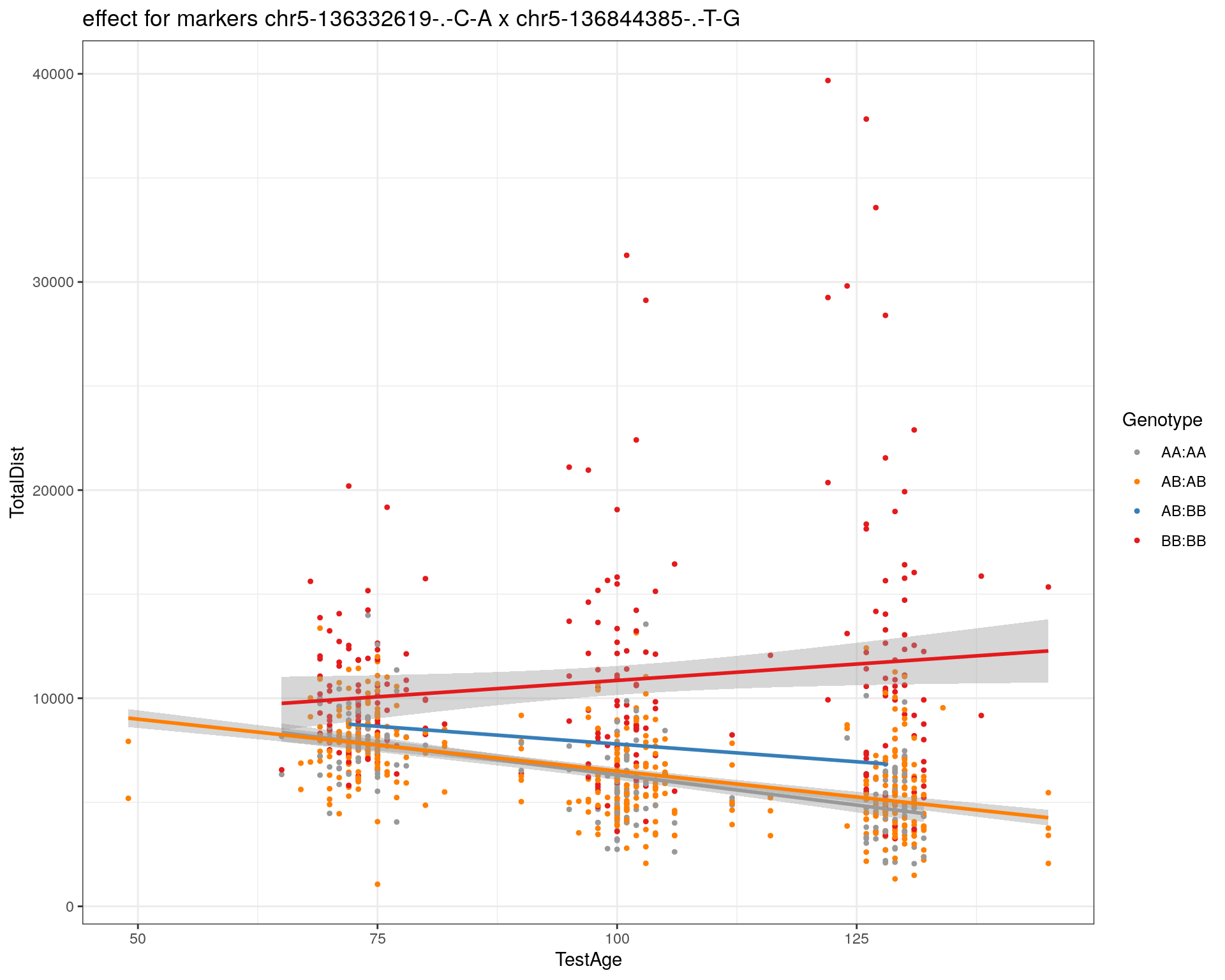
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
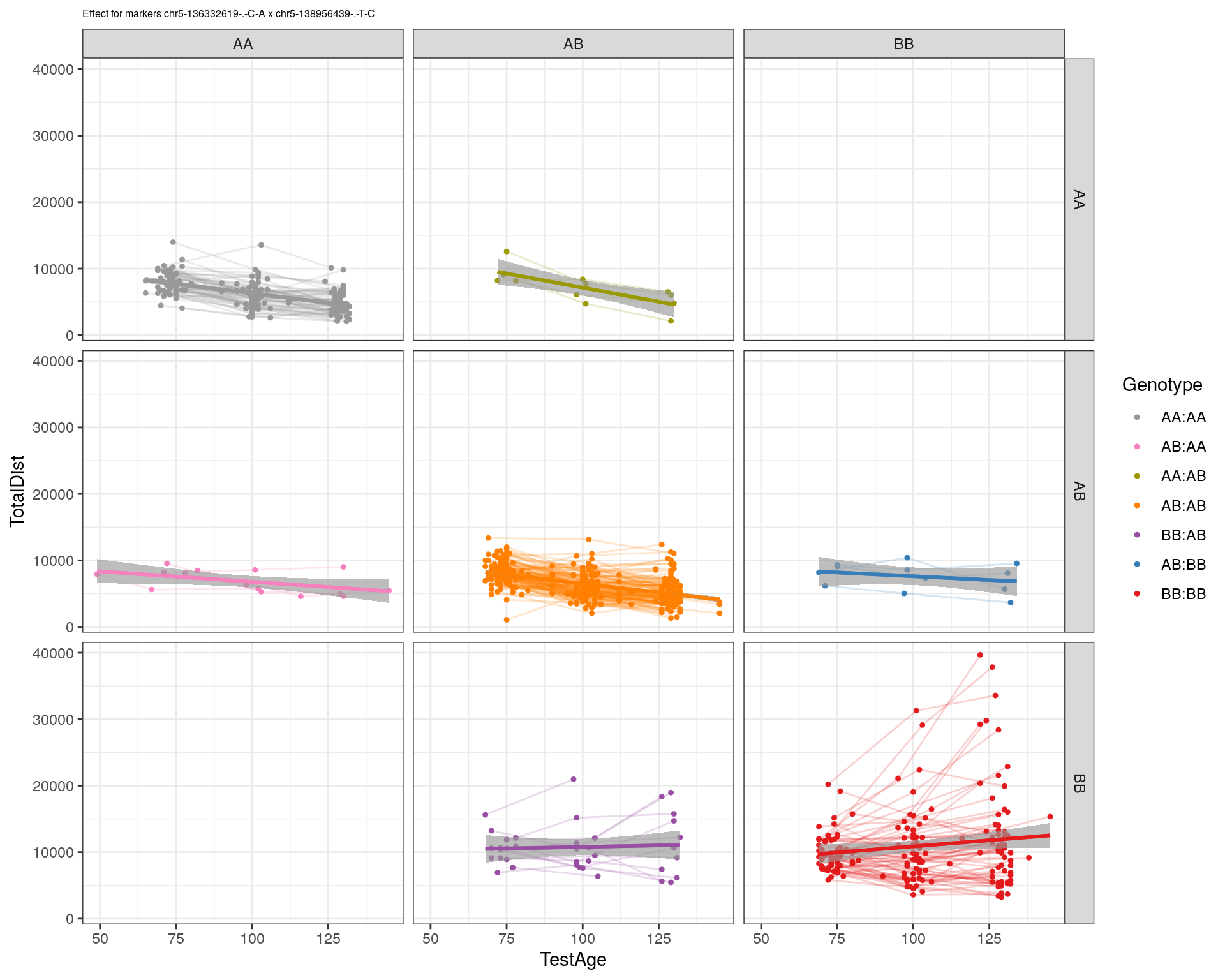
[1] "chr5-138956439-.-T-C"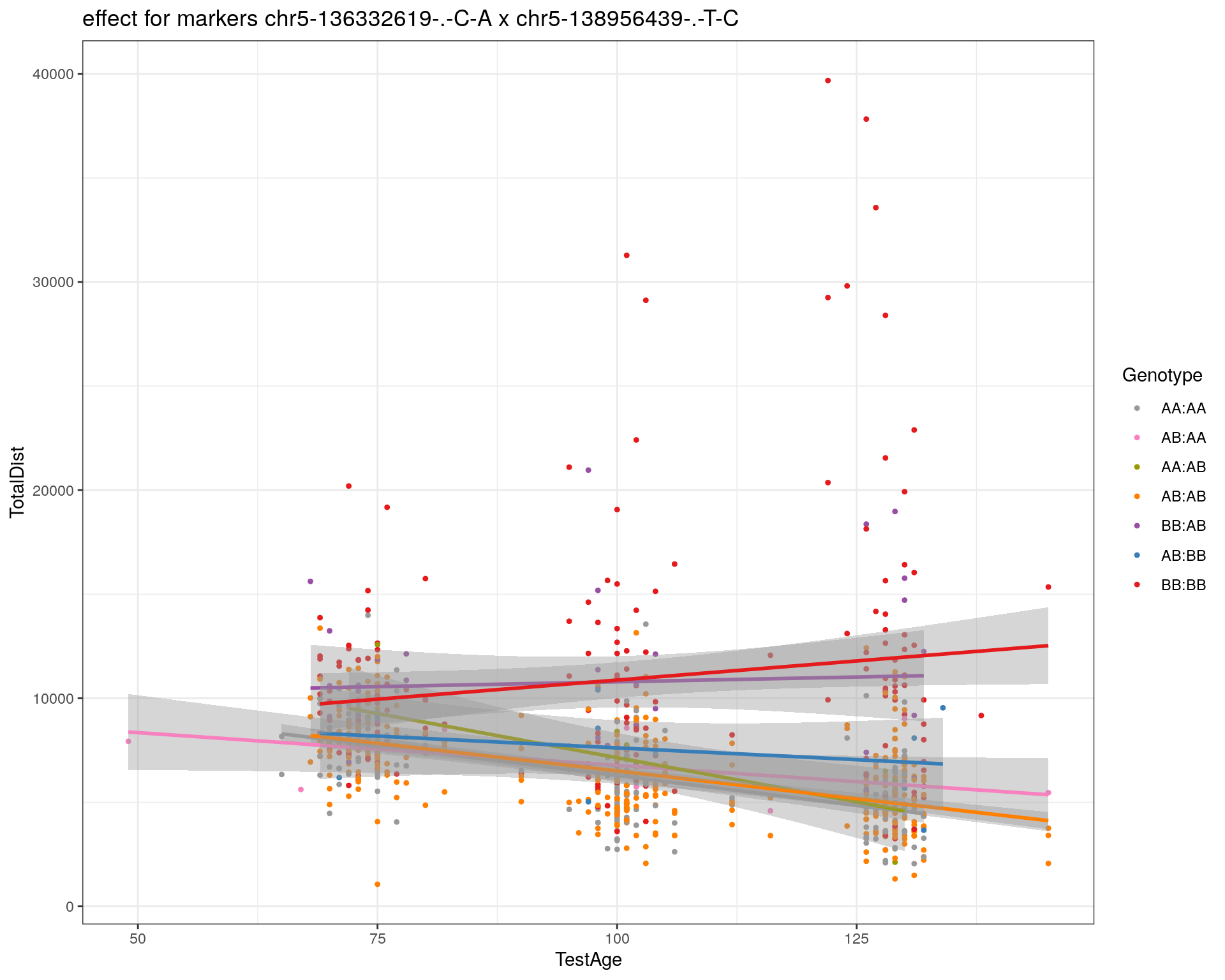
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
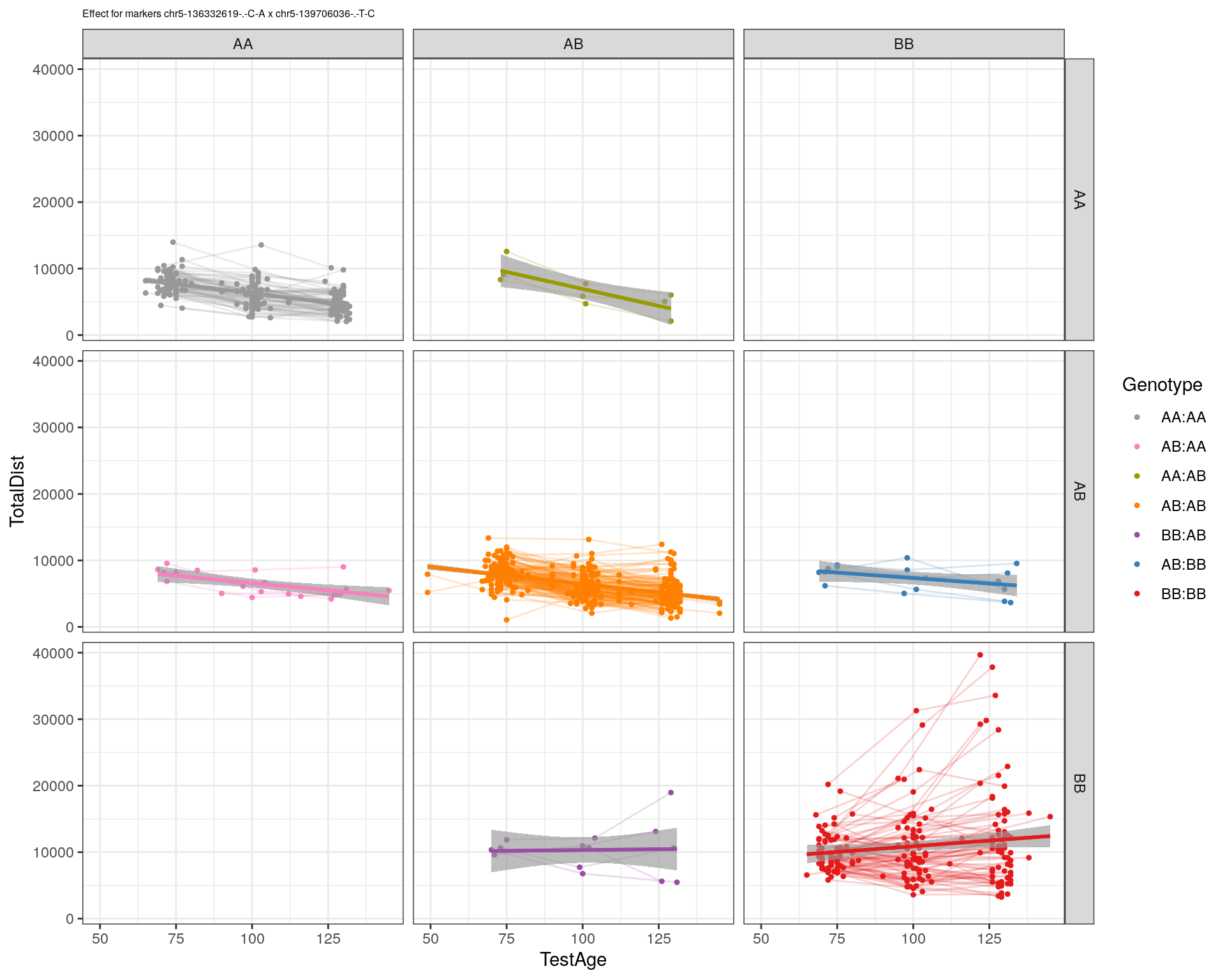
[1] "chr5-139706036-.-T-C"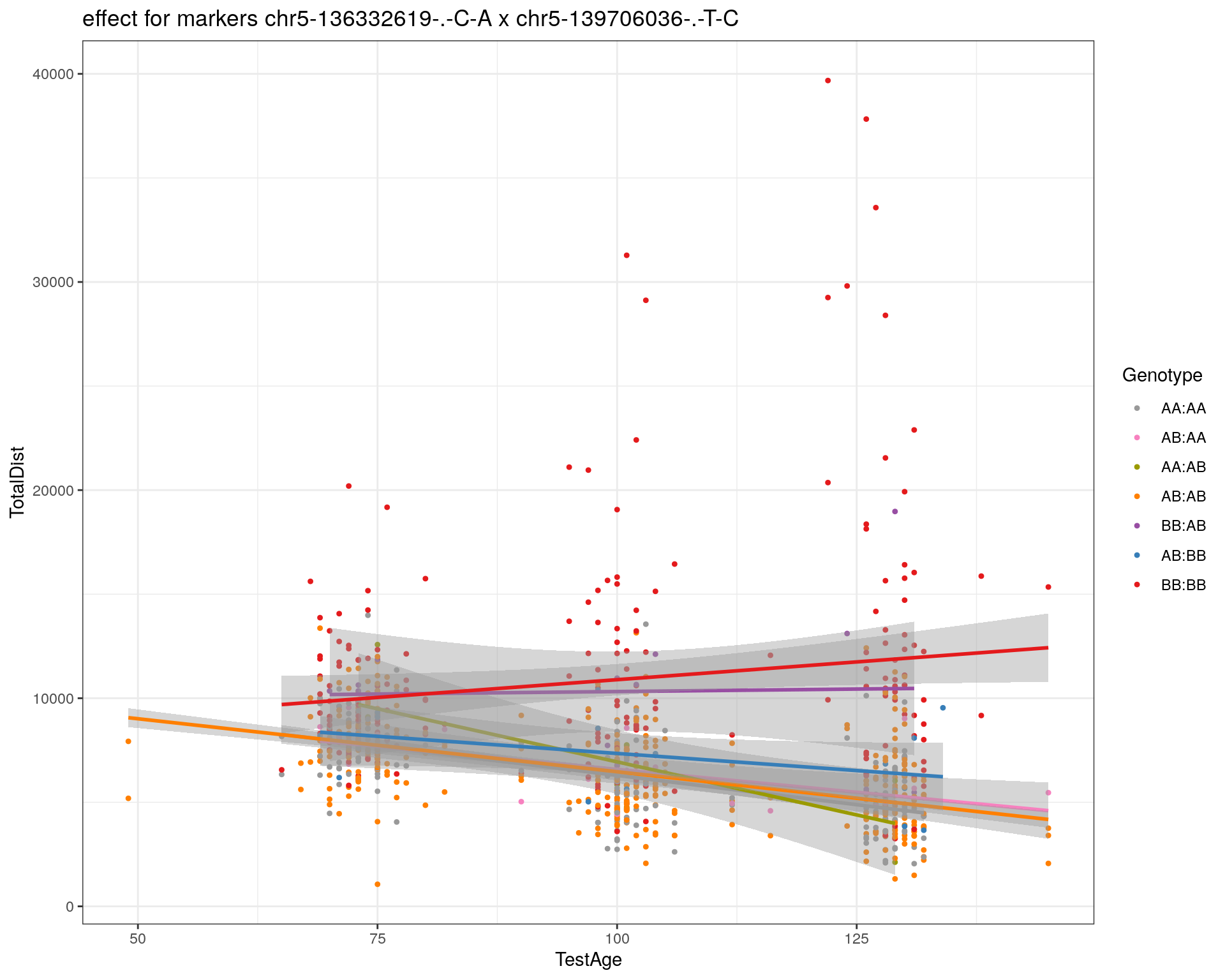
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
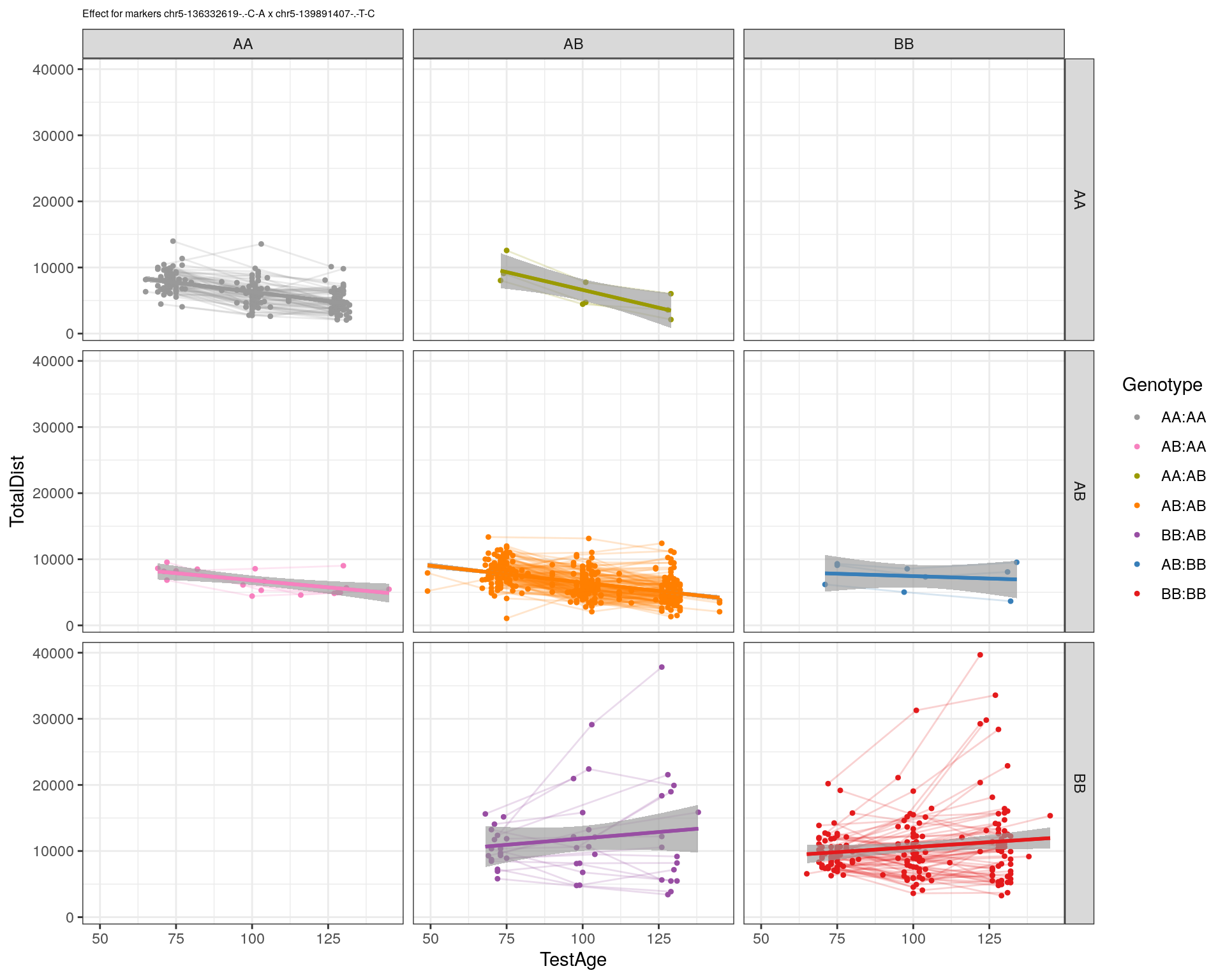
[1] "chr5-139891407-.-T-C"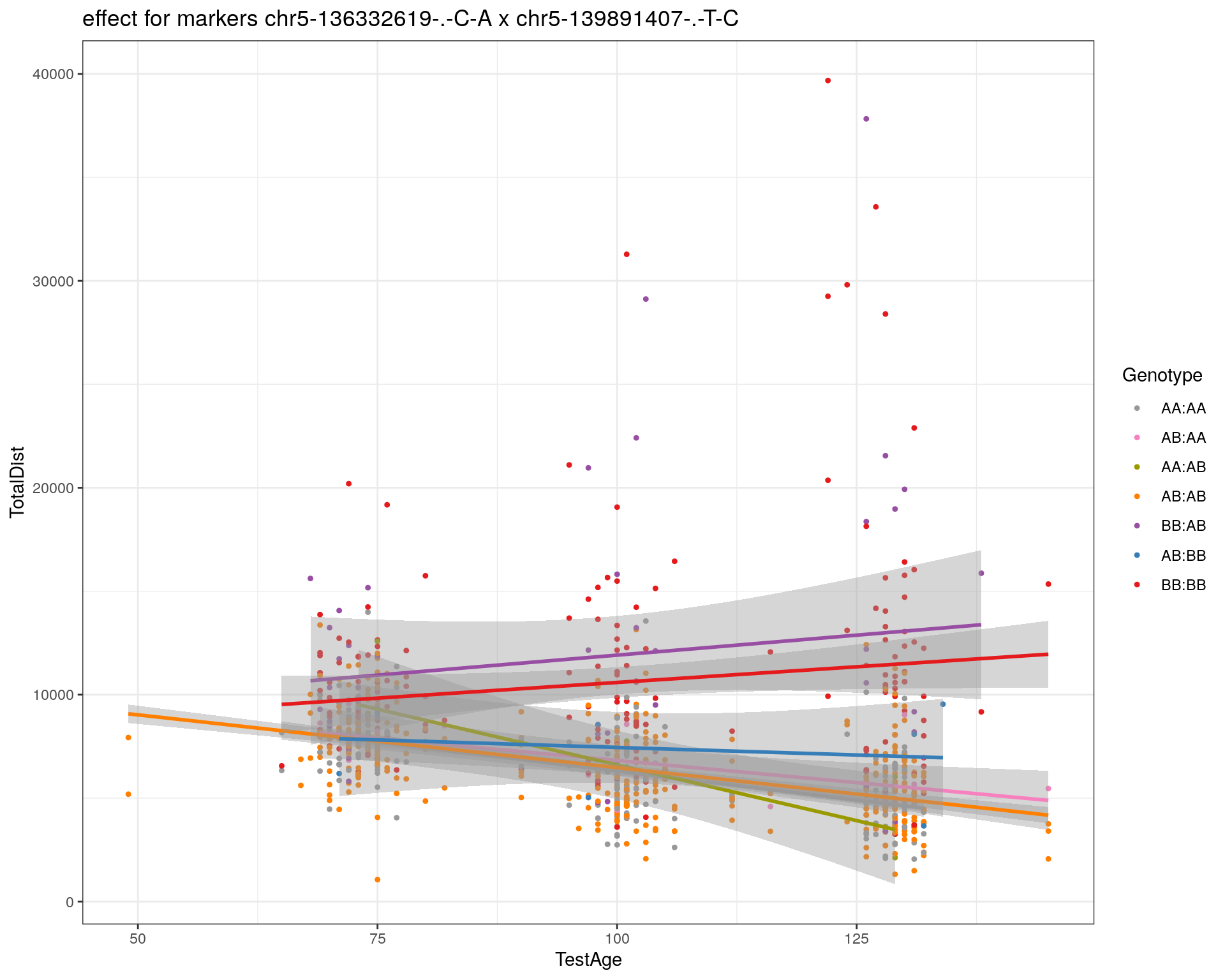
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr5-140071081-.-C-A"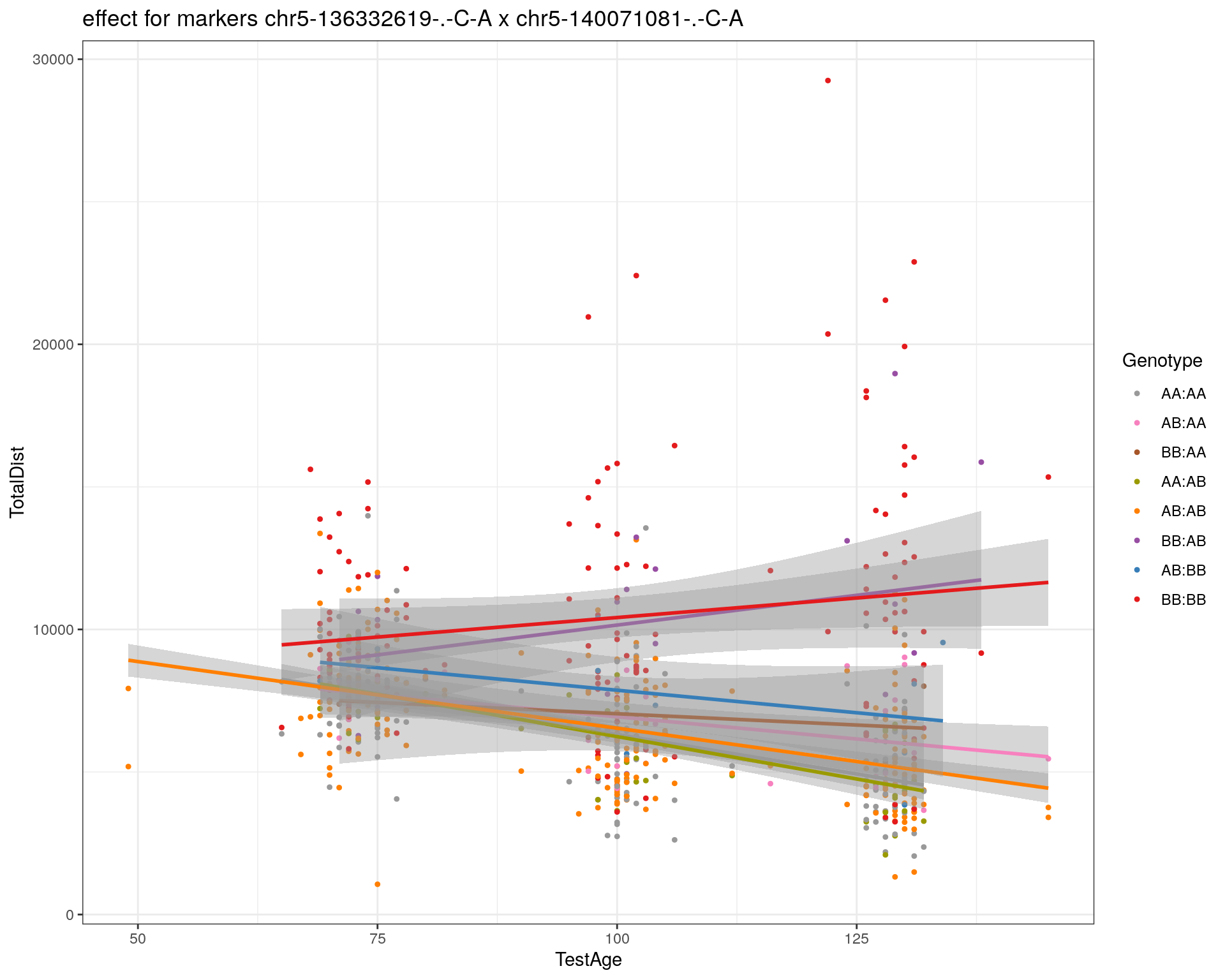
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
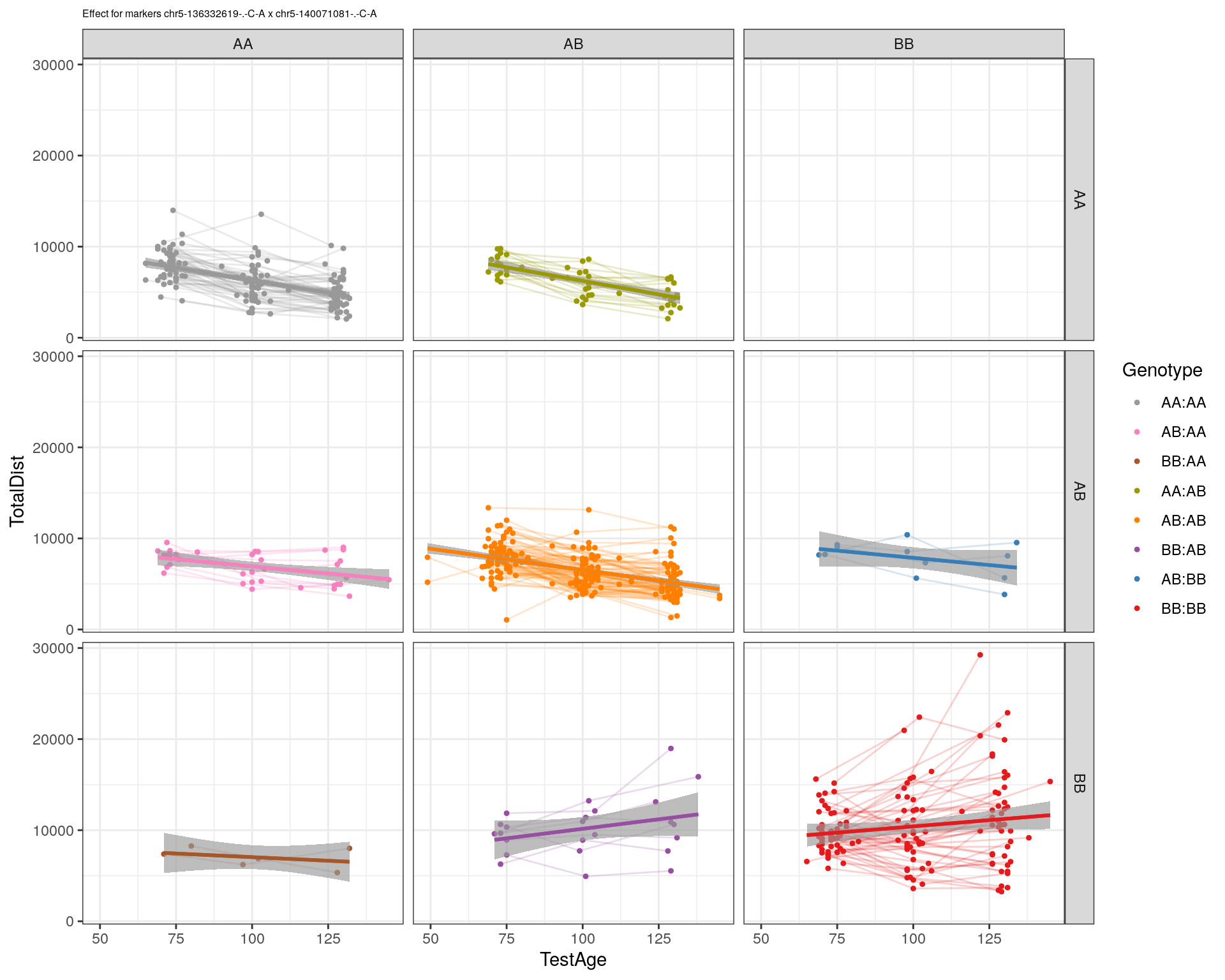
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr7-35061547-.-A-G"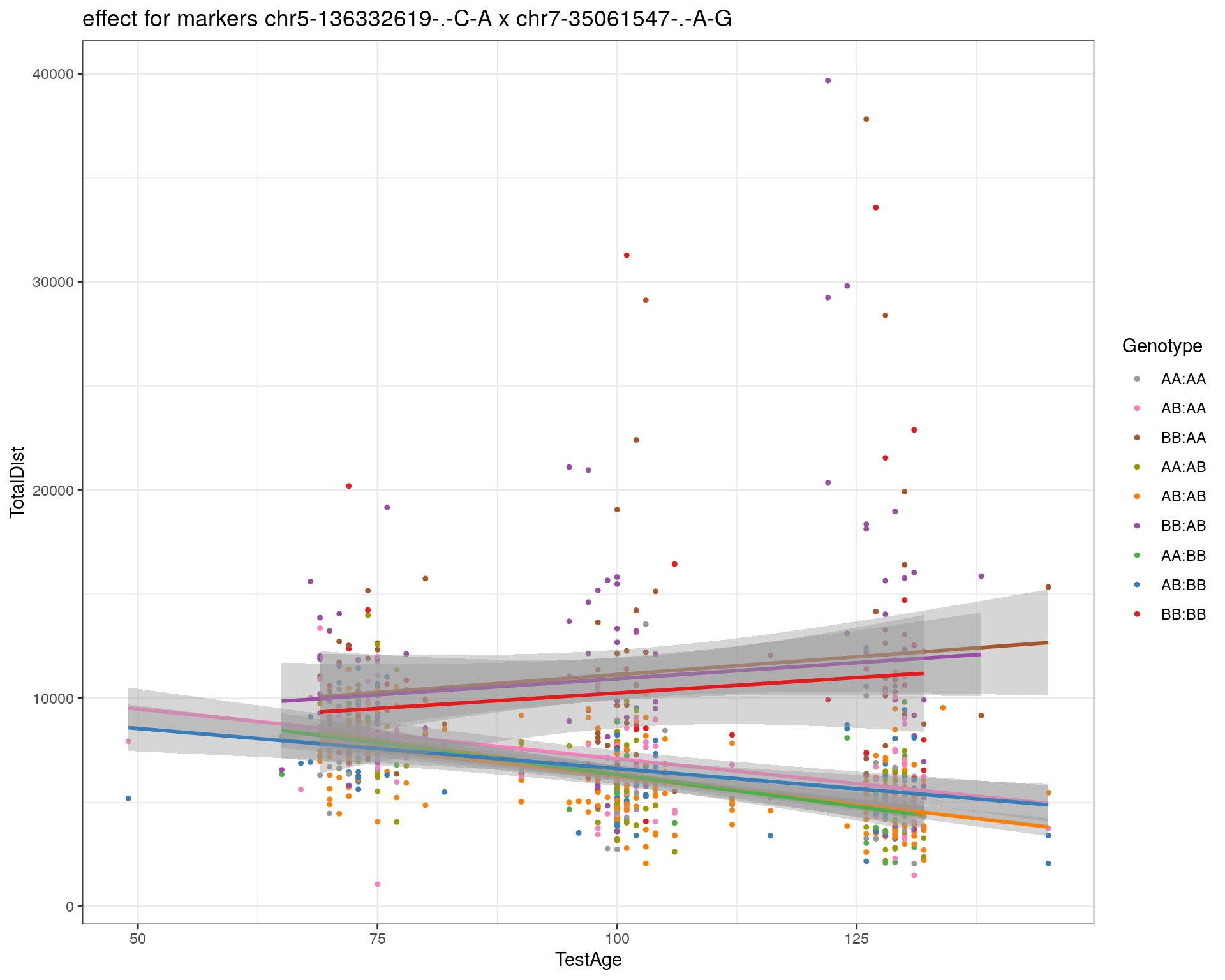
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
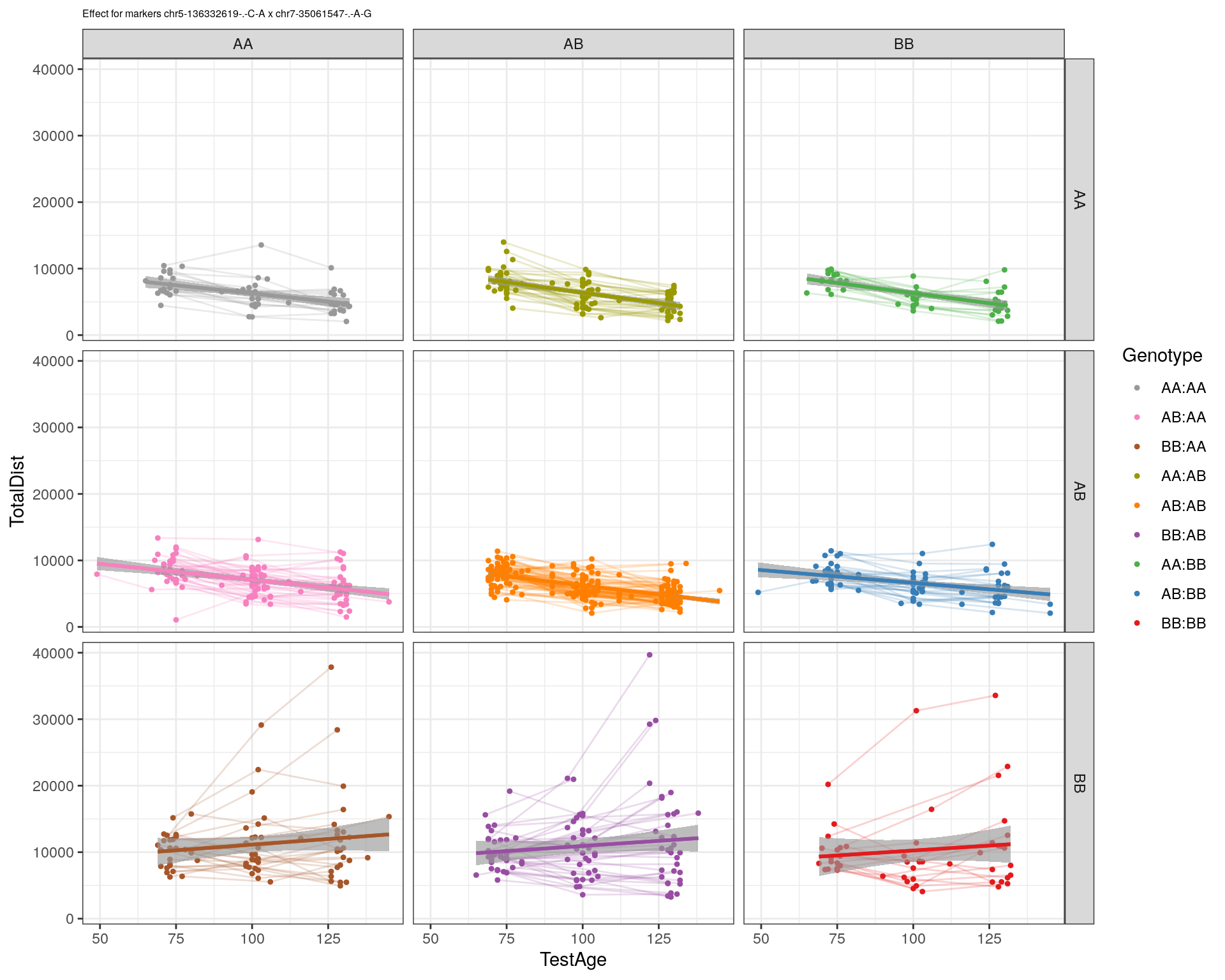
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr7-44272713-.-T-C"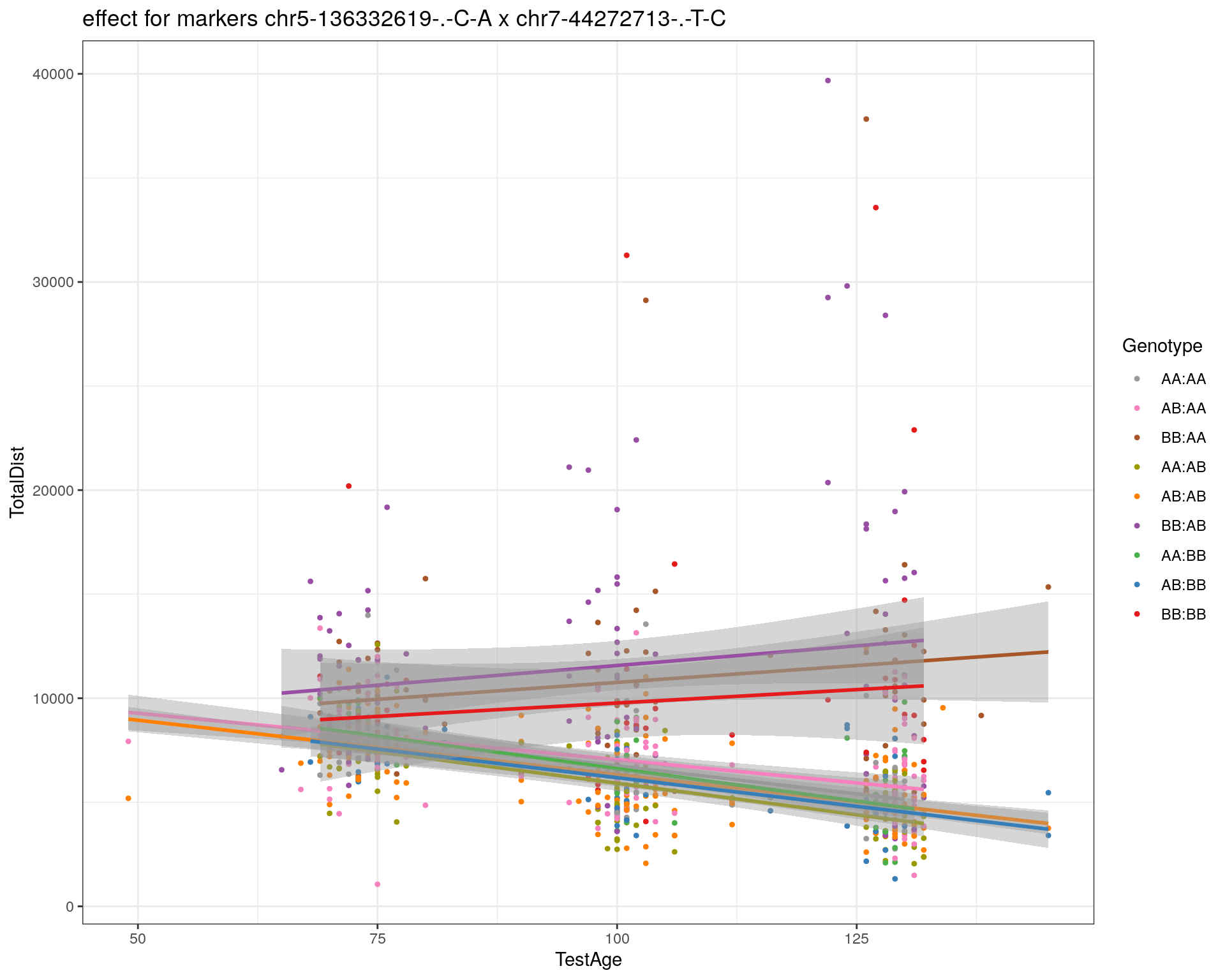
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
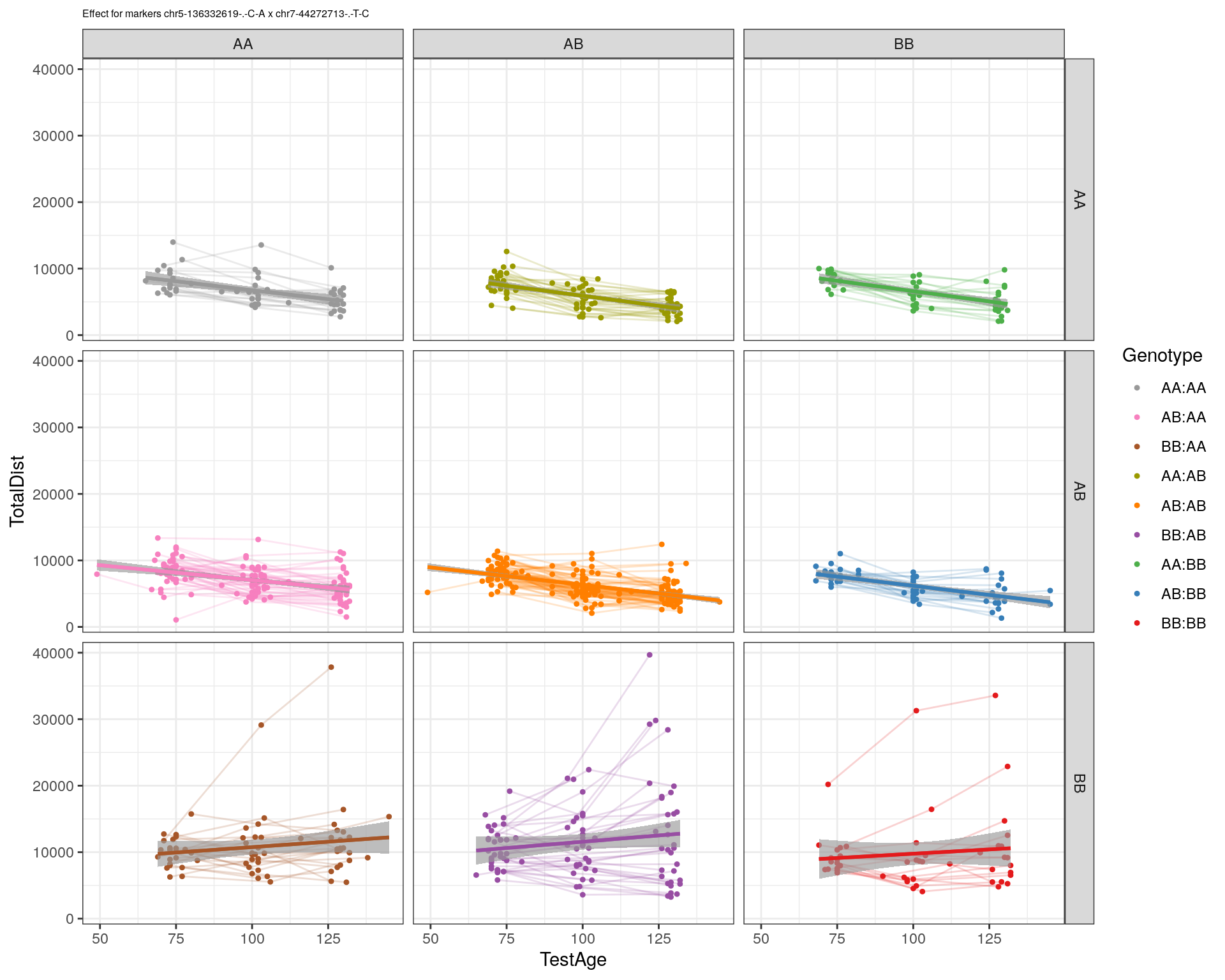
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr7-66785411-.-A-G"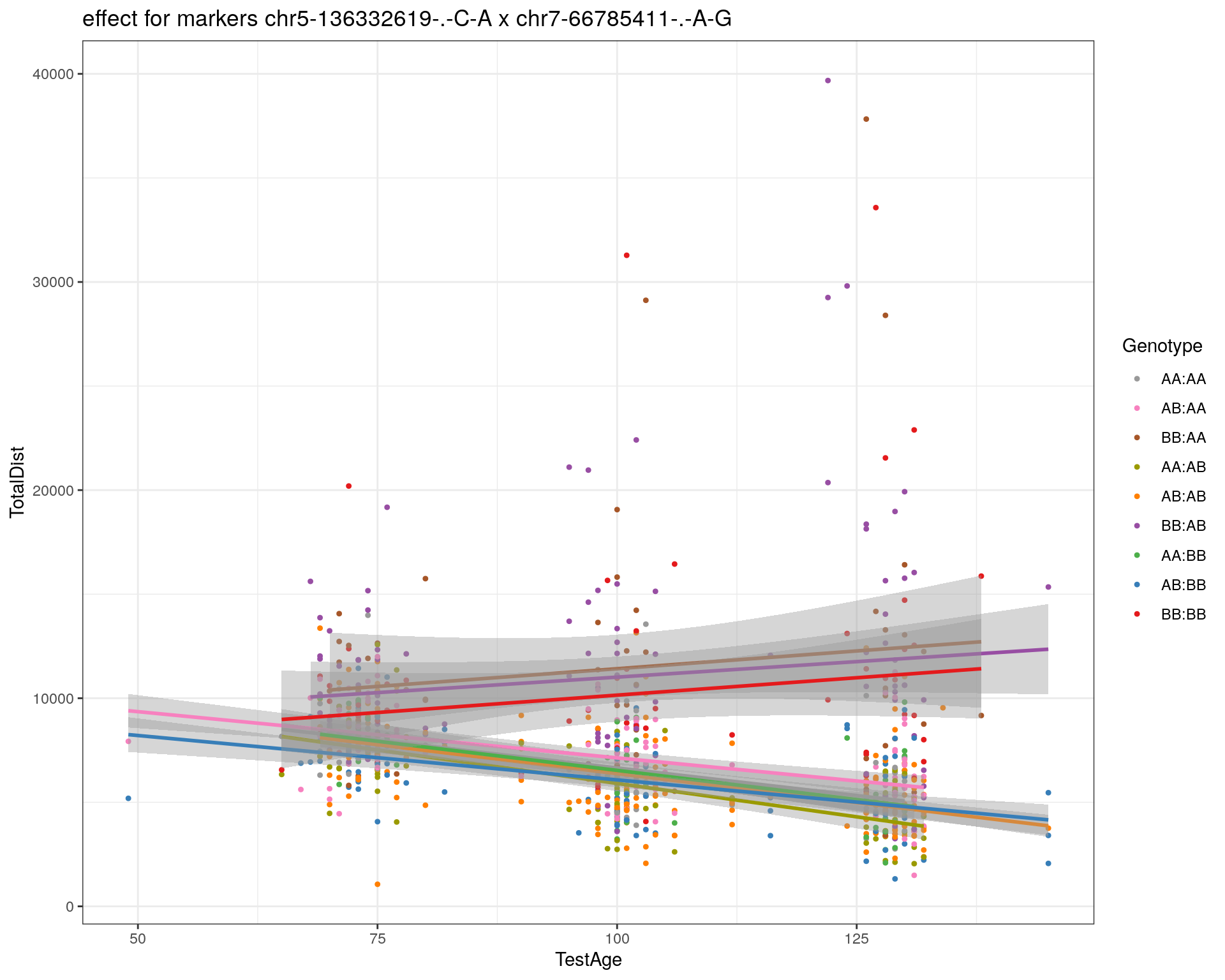
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
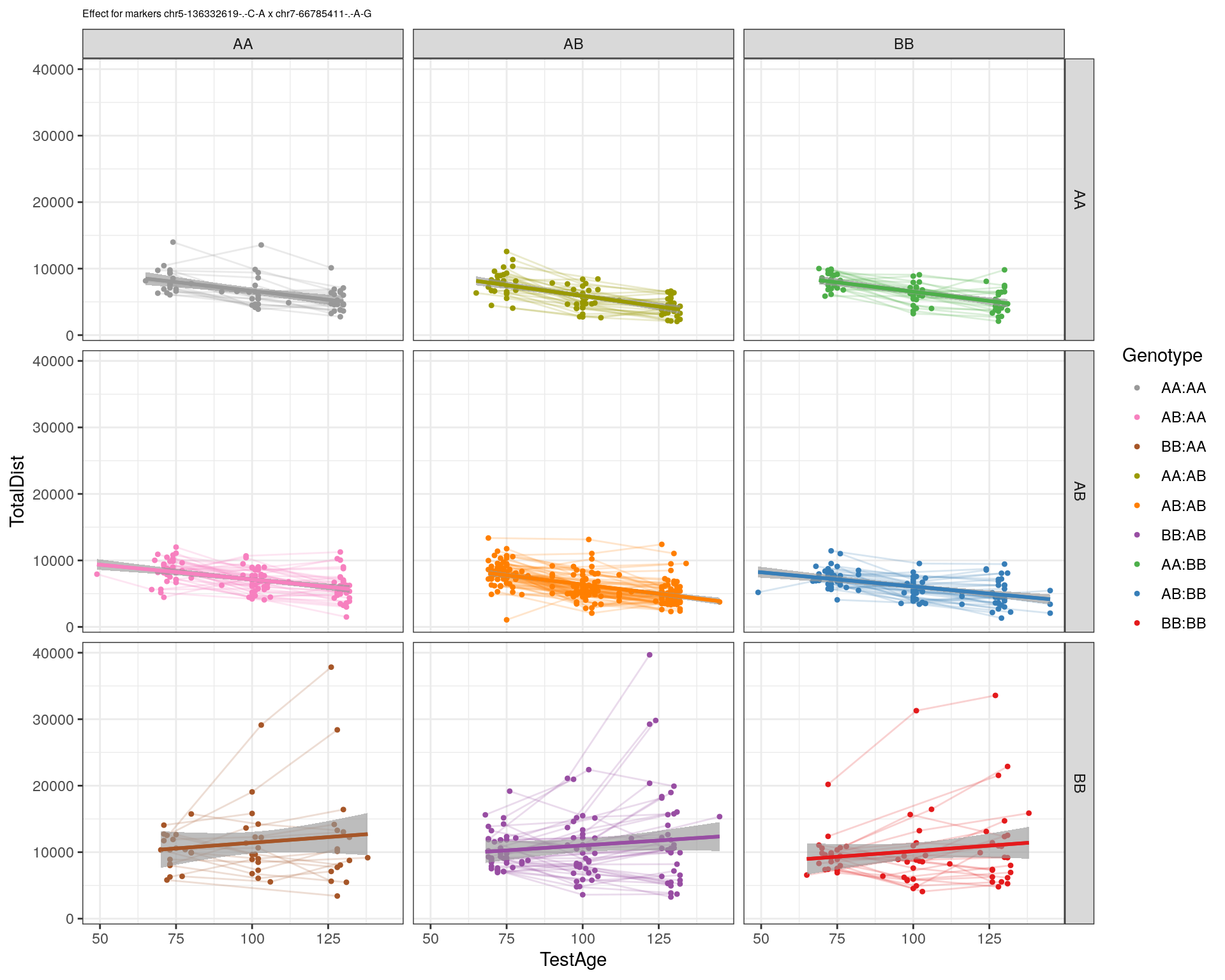
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr7-112510032-.-A-T"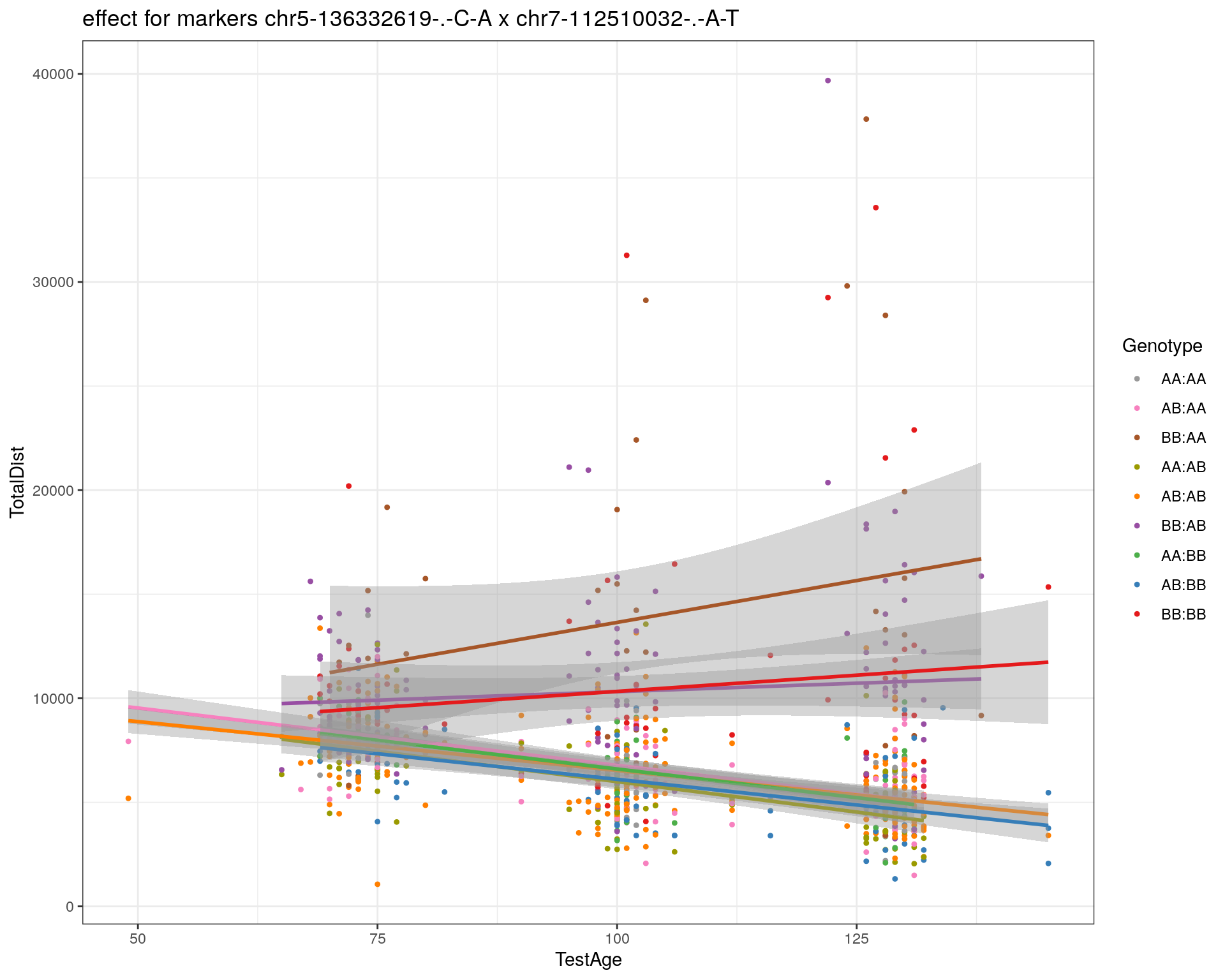
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
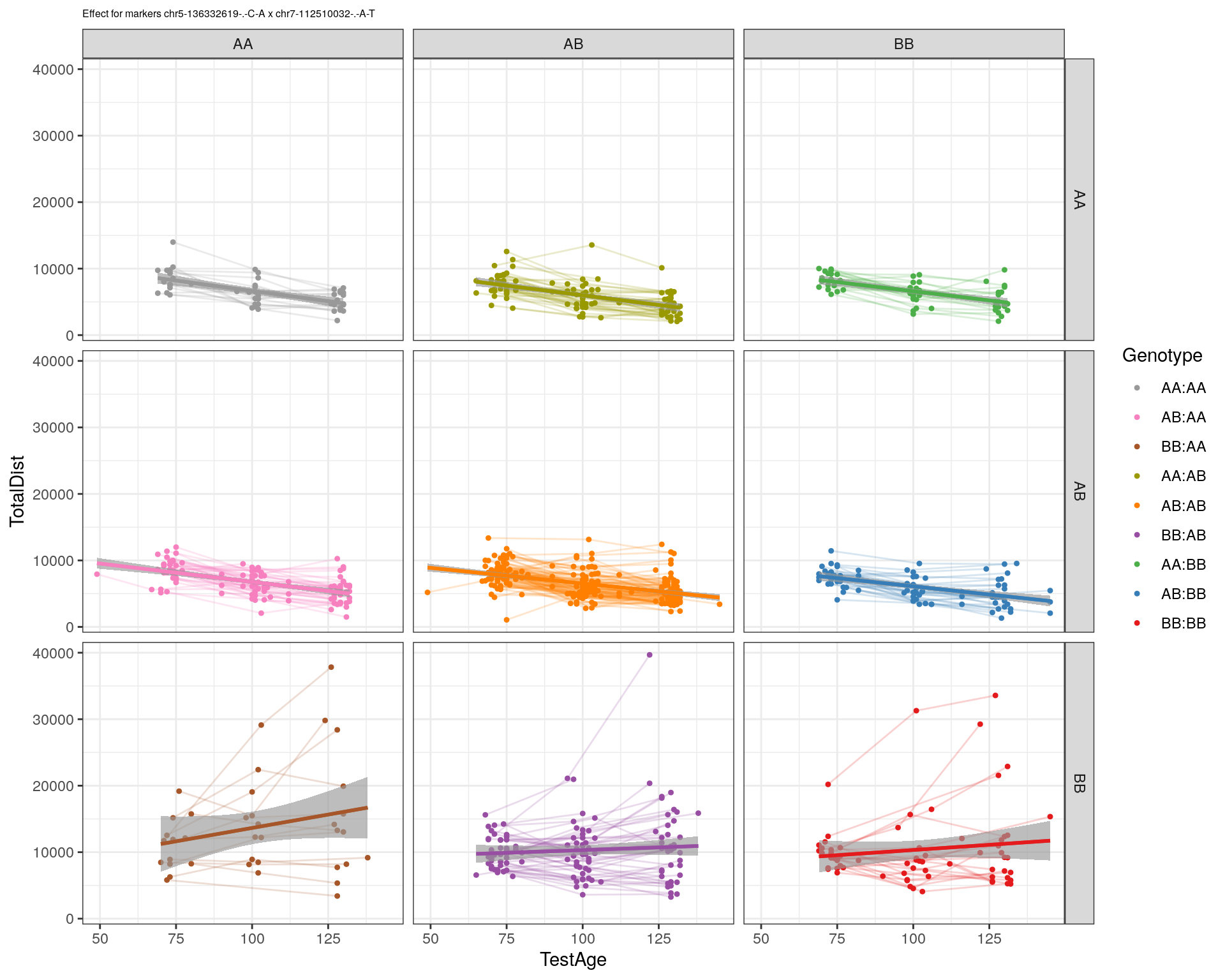
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr8-91986173-.-T-C"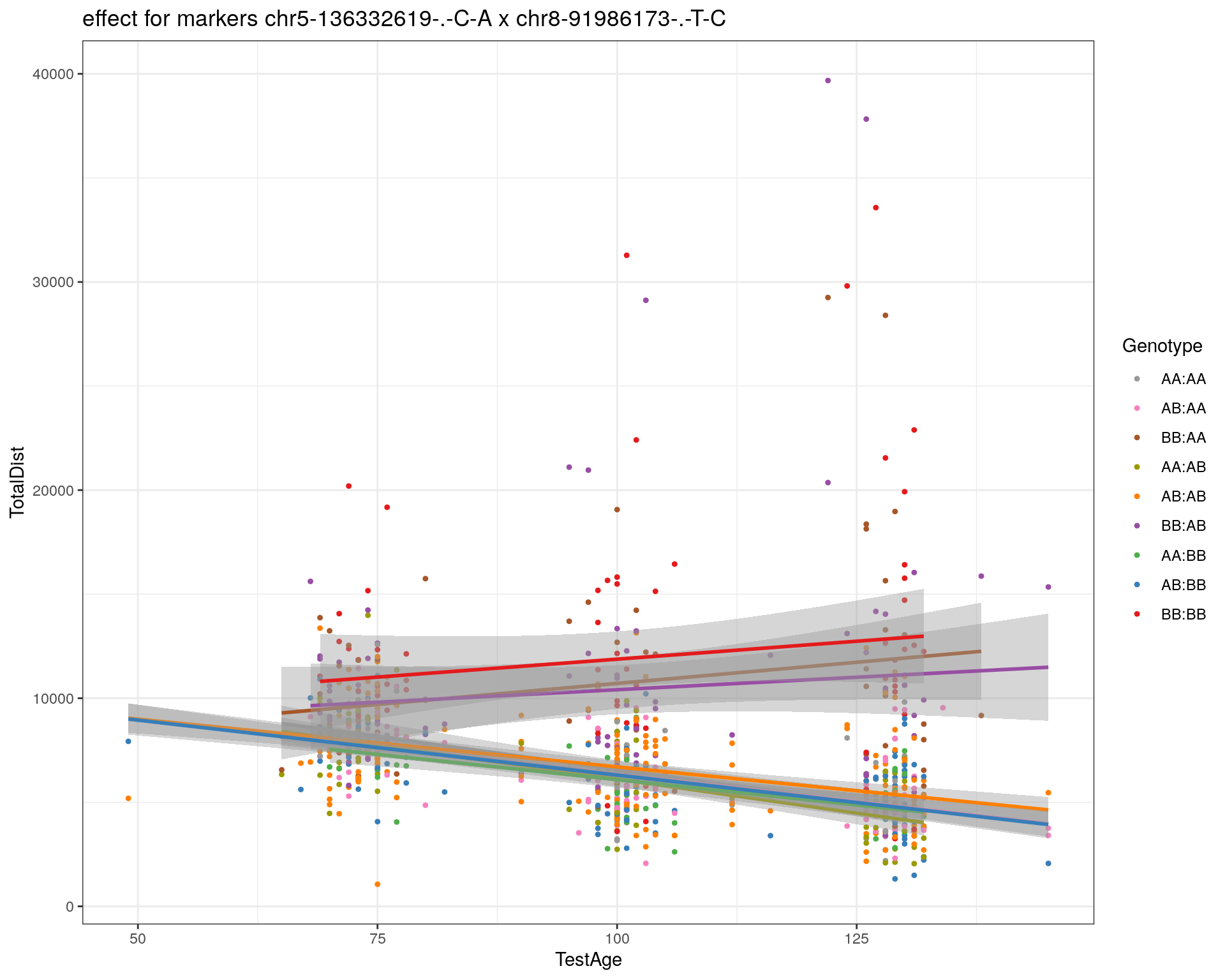
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
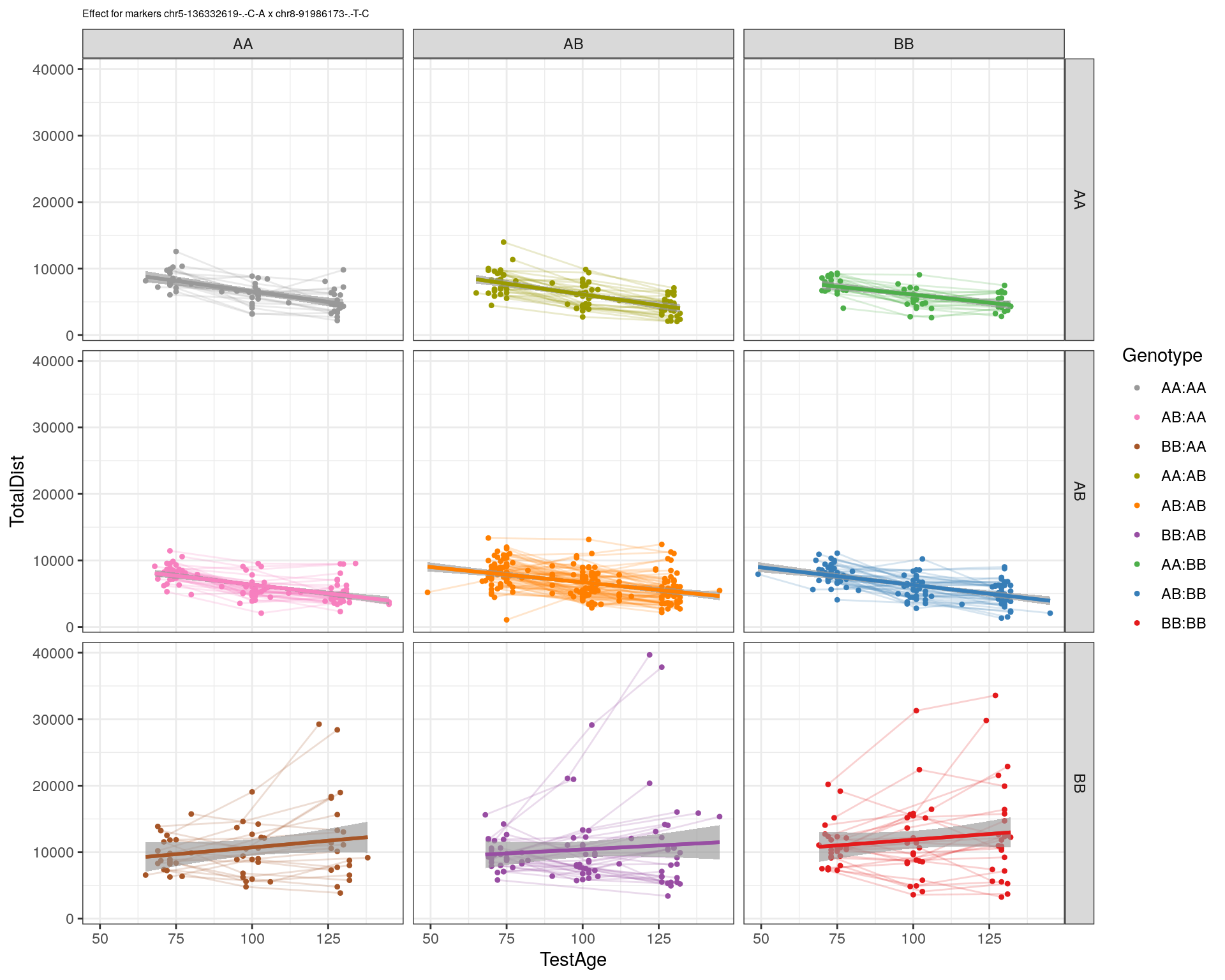
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr8-99395759-.-T-C"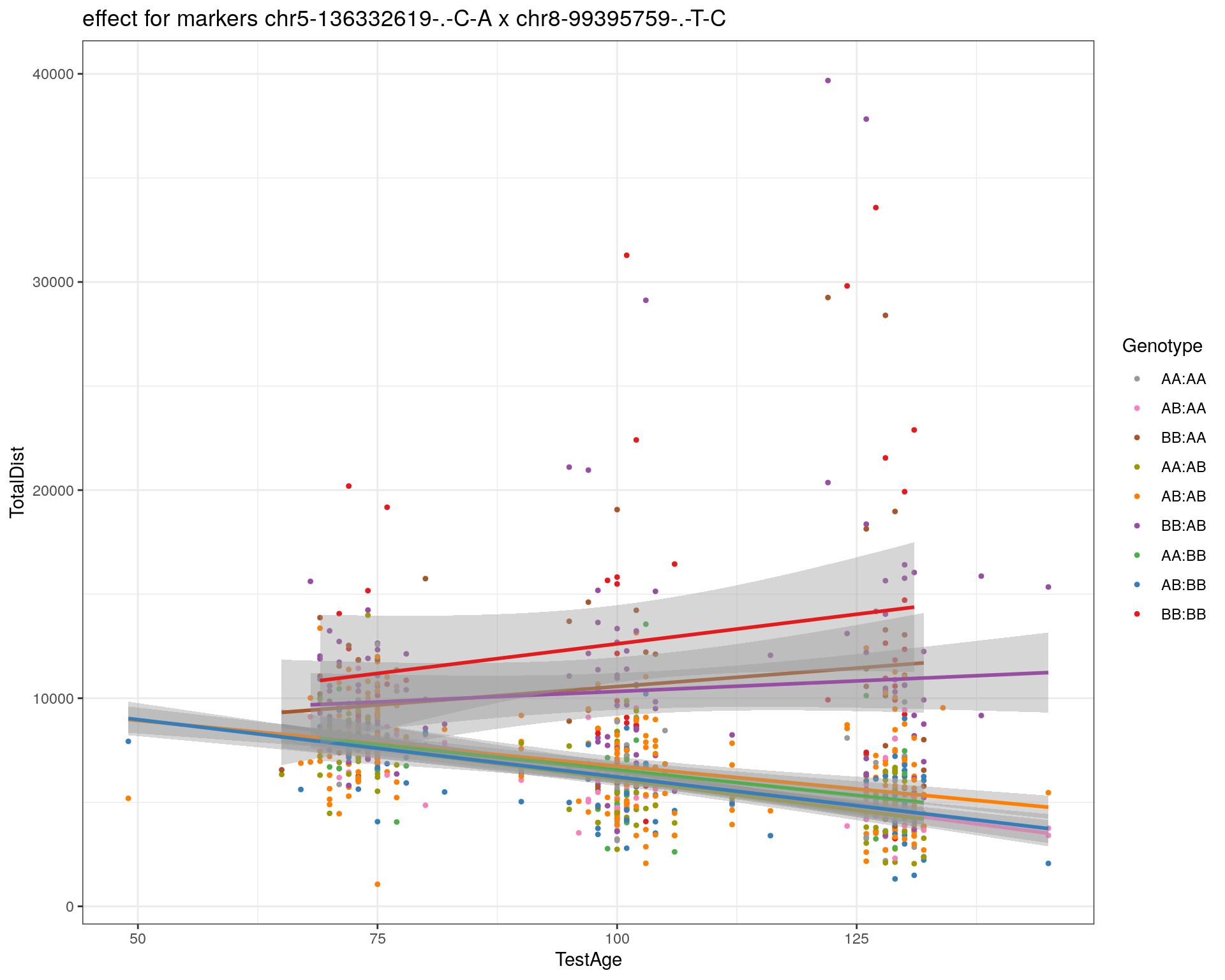
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
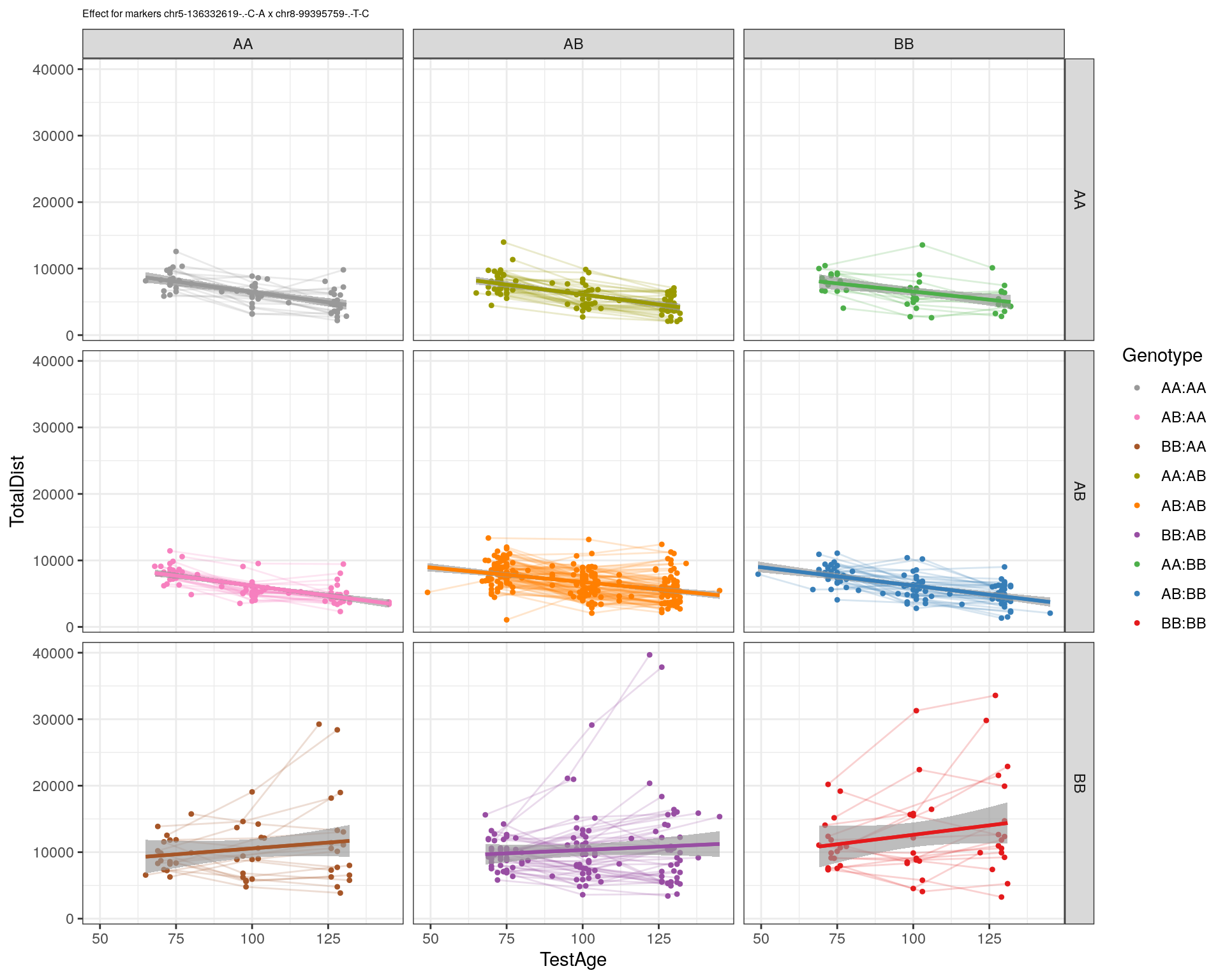
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr8-112465004-.-G-A"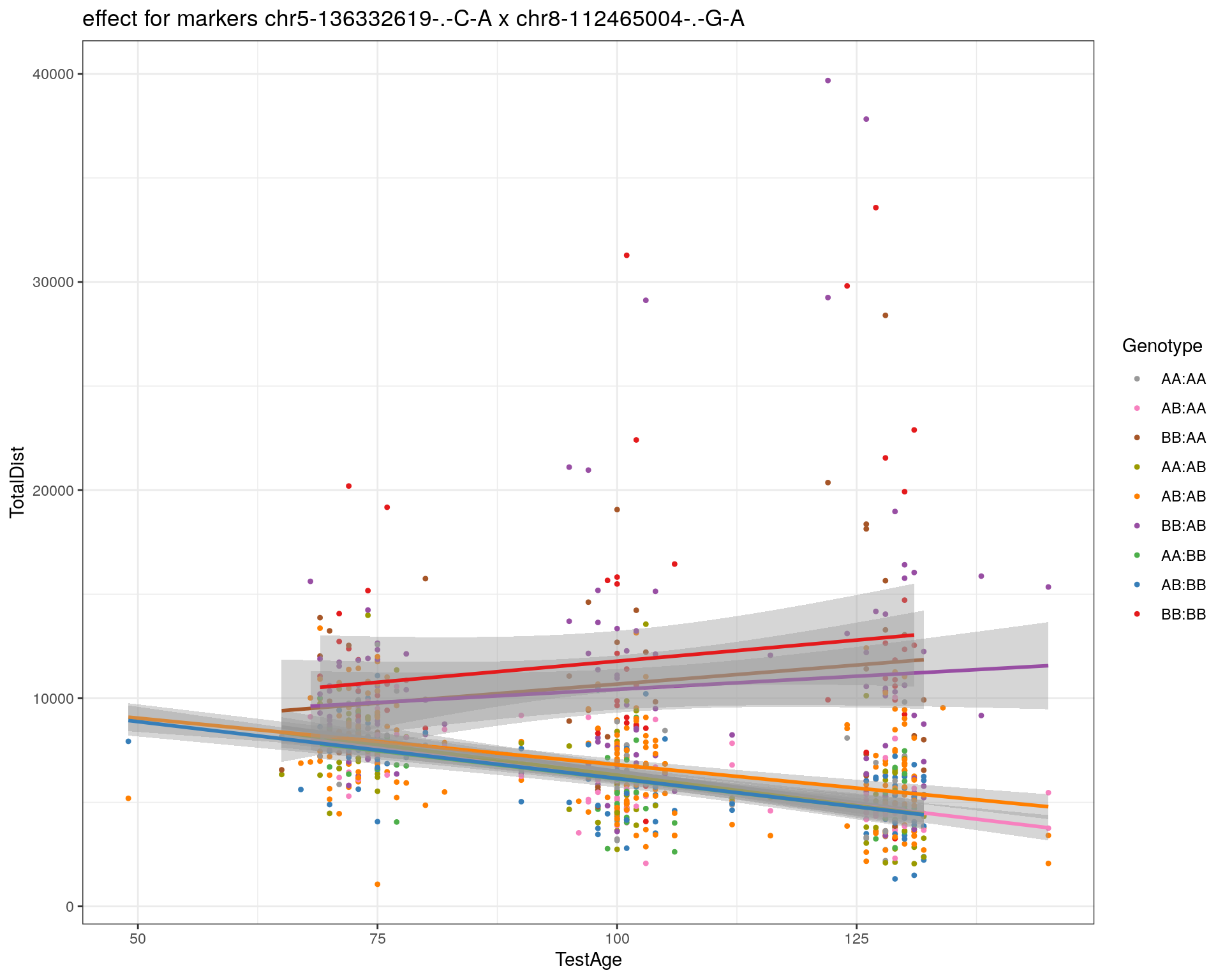
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
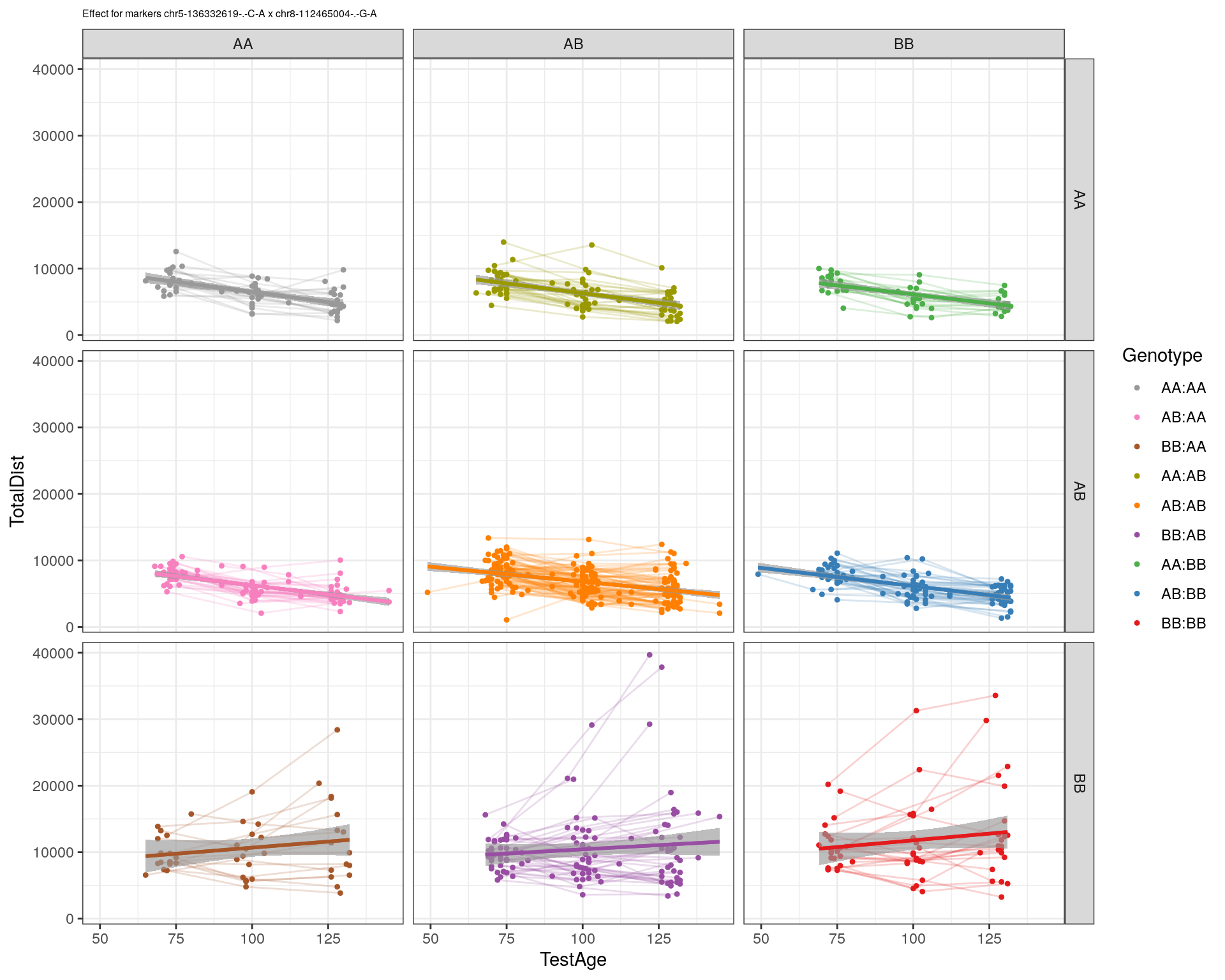
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr9-64698947-.-C-T"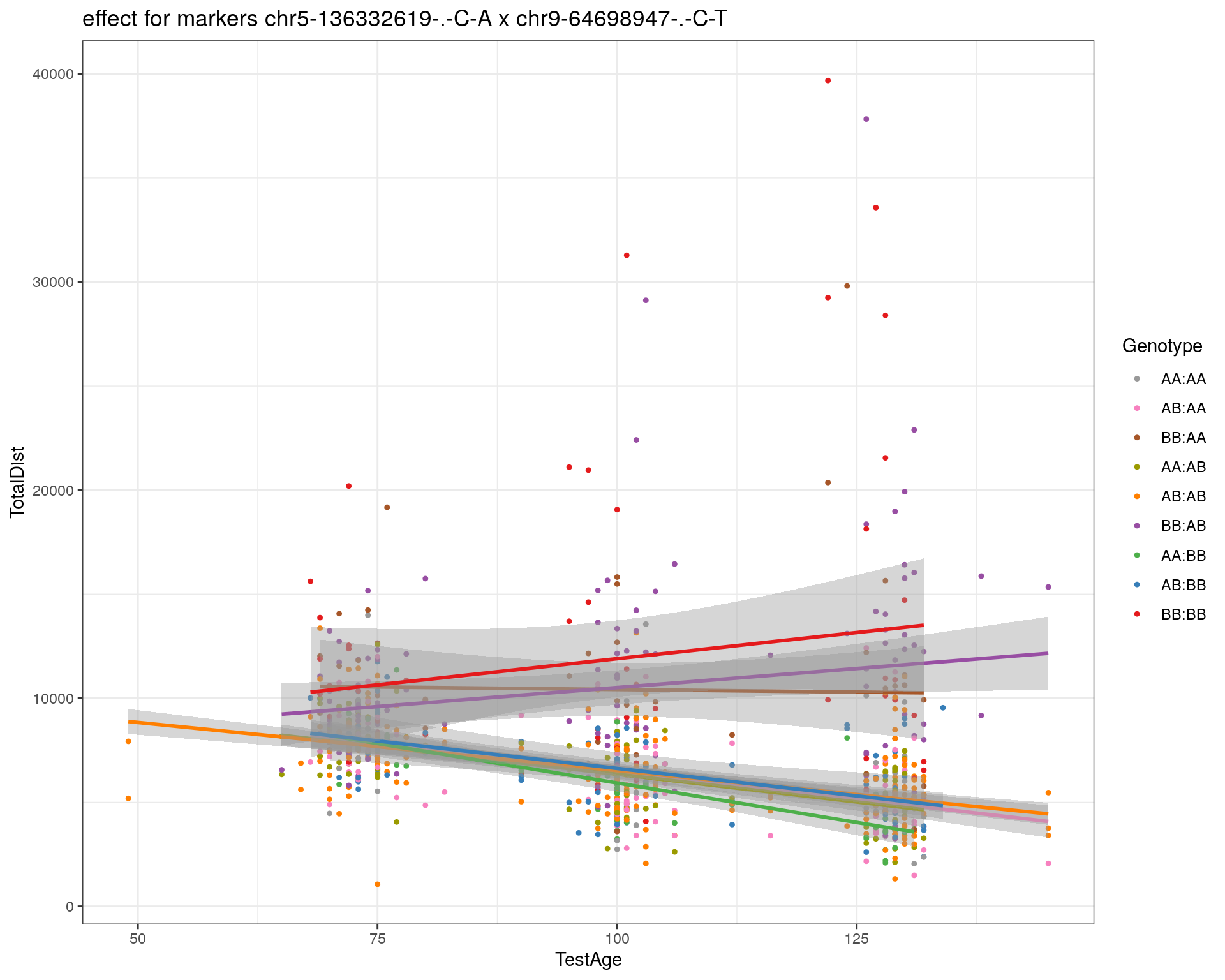
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
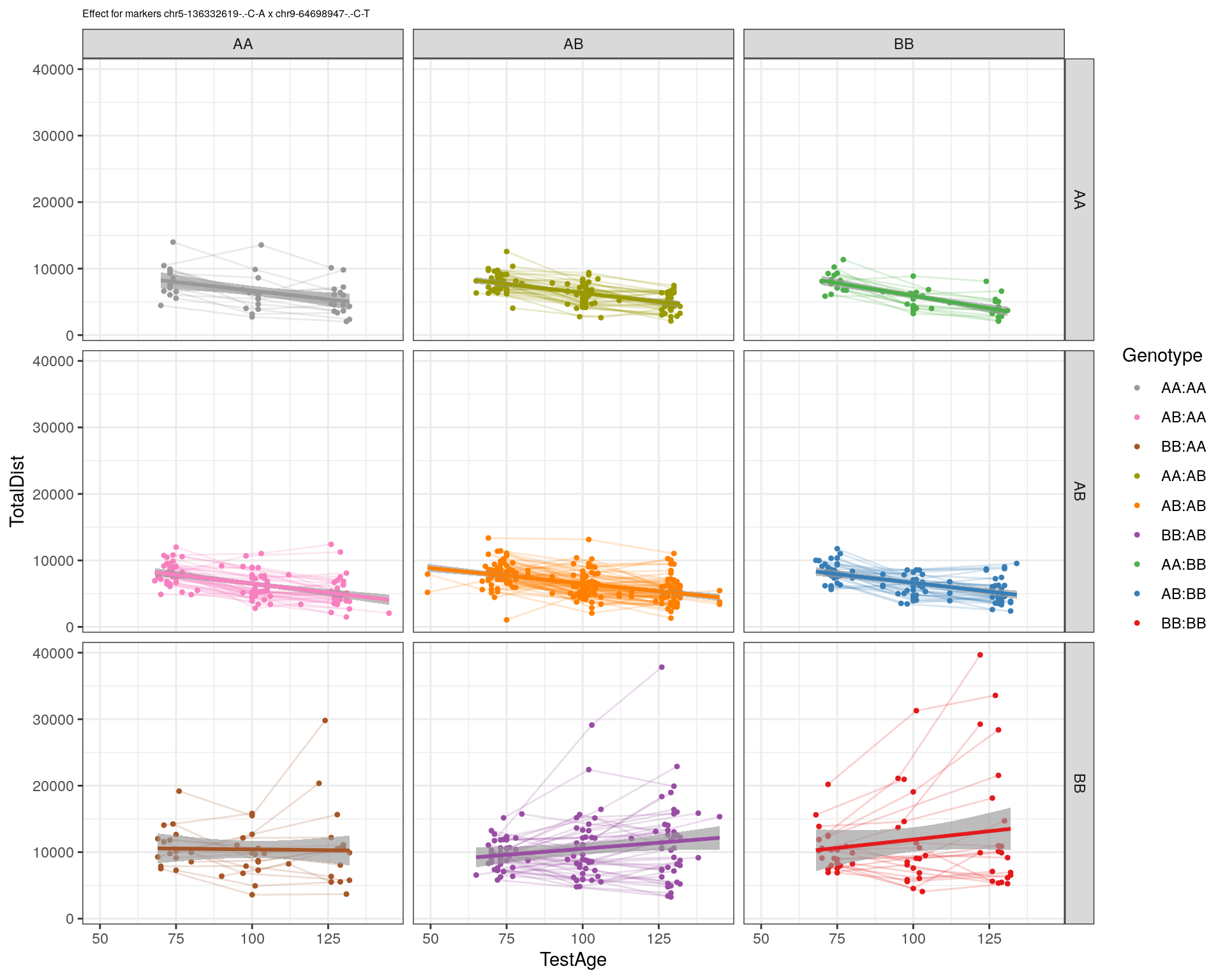
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr9-65857475-.-T-C"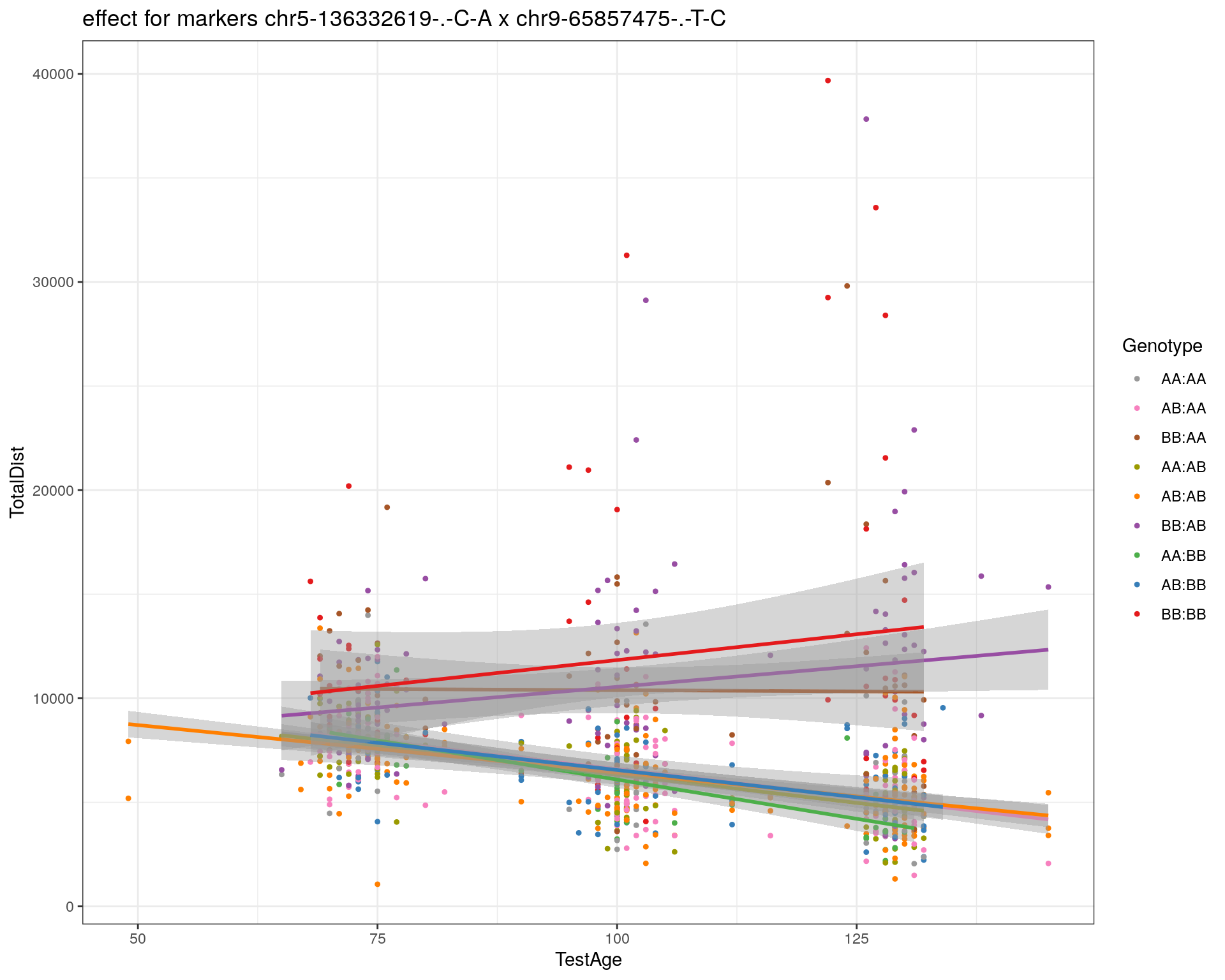
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
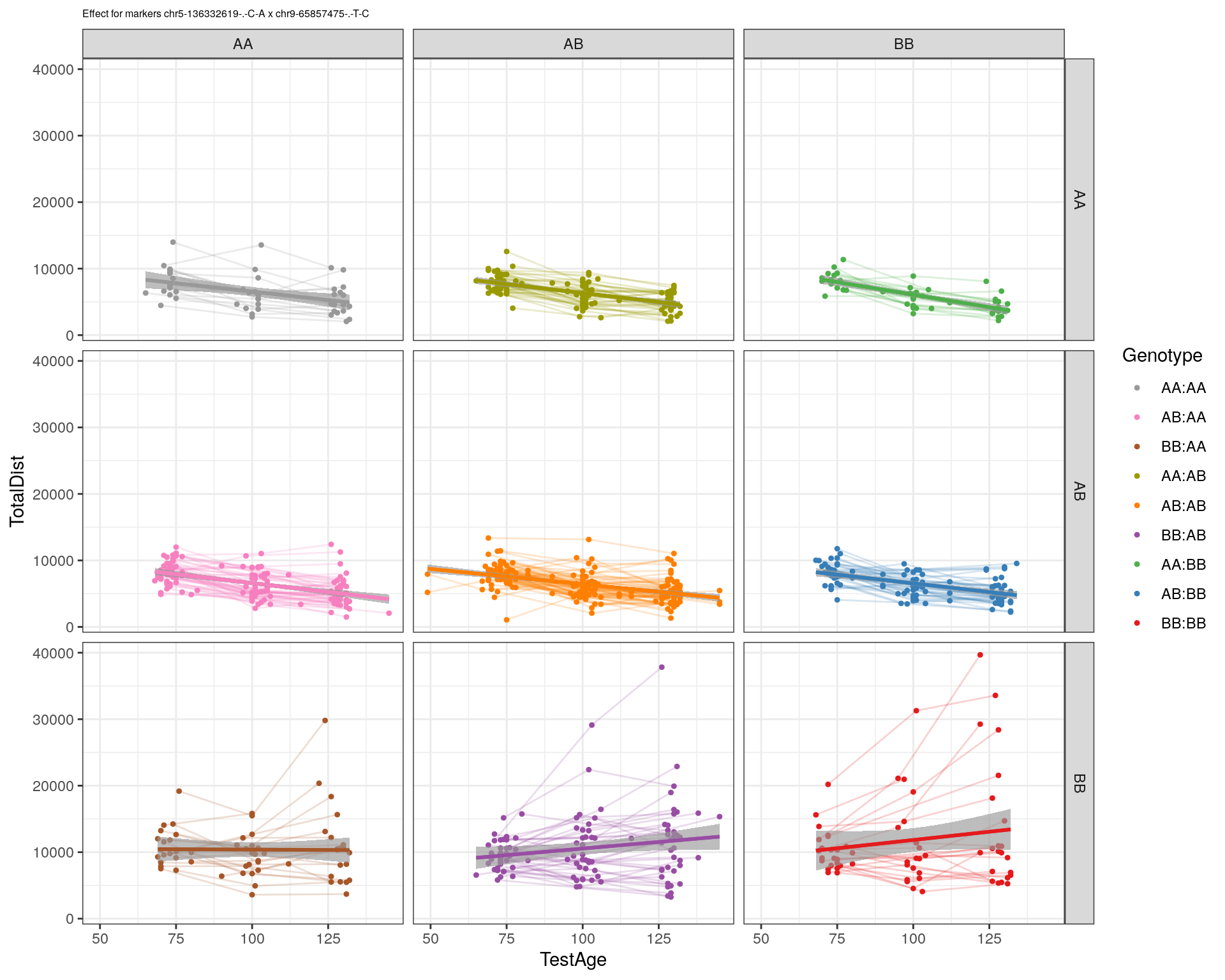
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr9-69415567-.-T-A"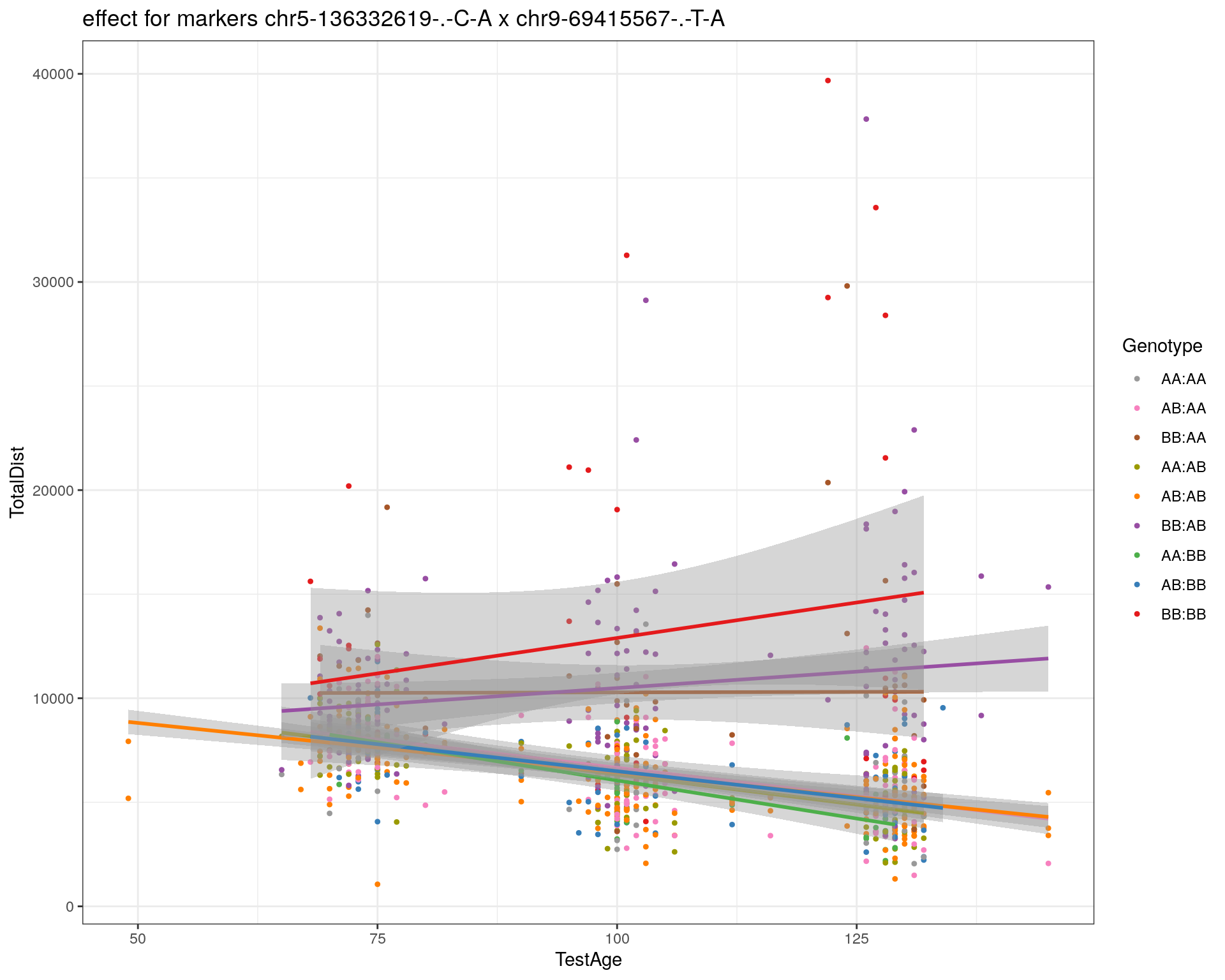
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
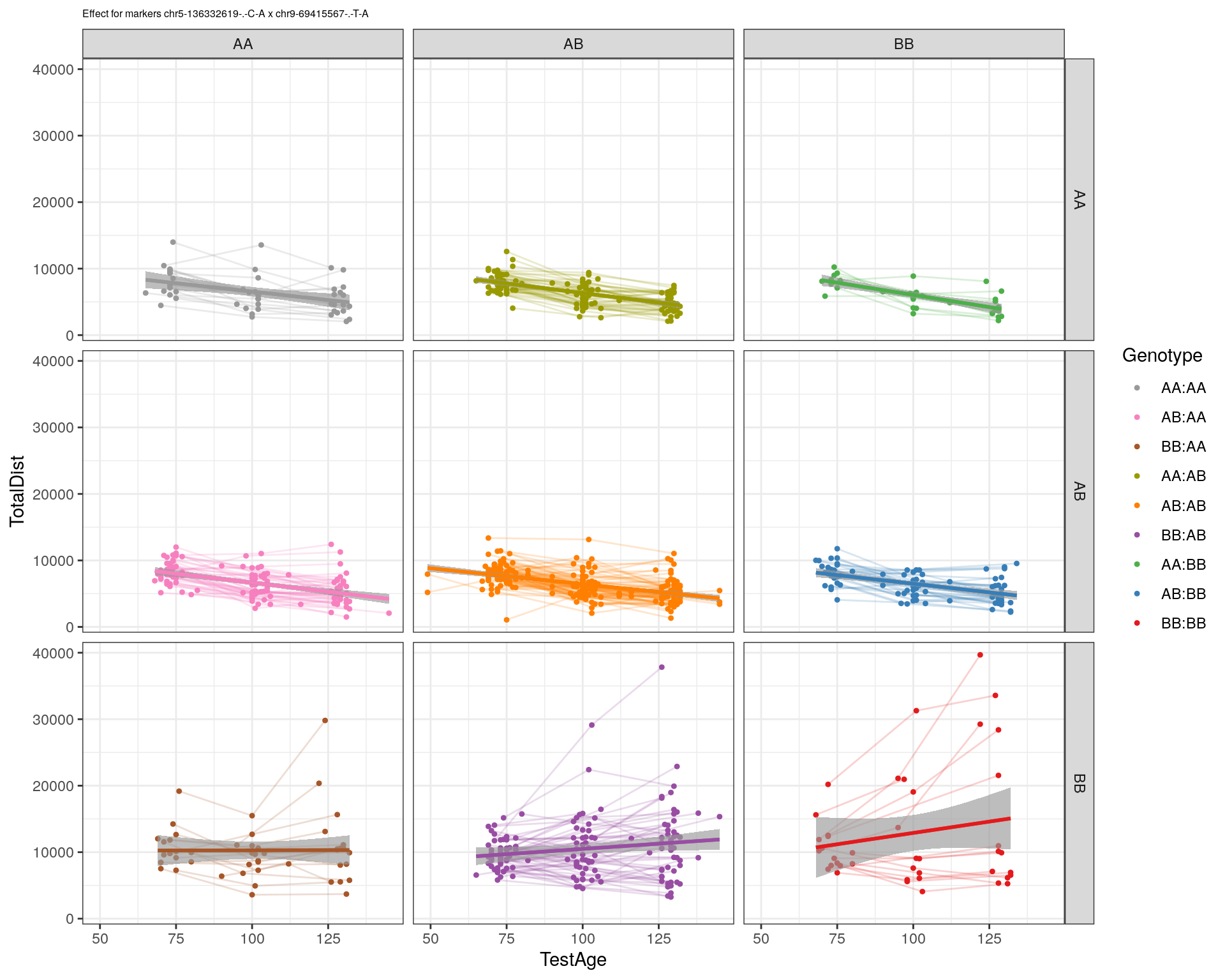
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr9-71739608-.-T-A"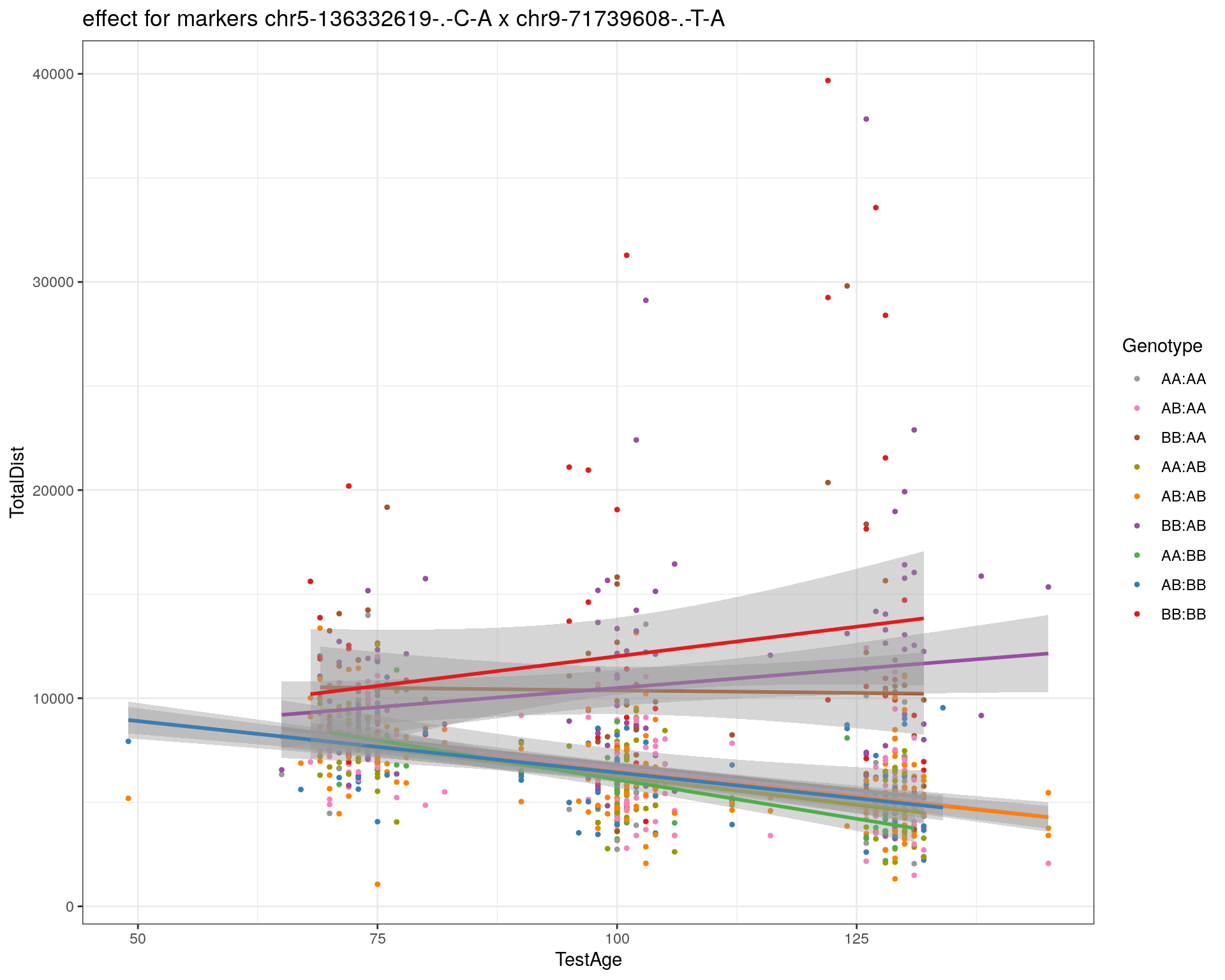
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
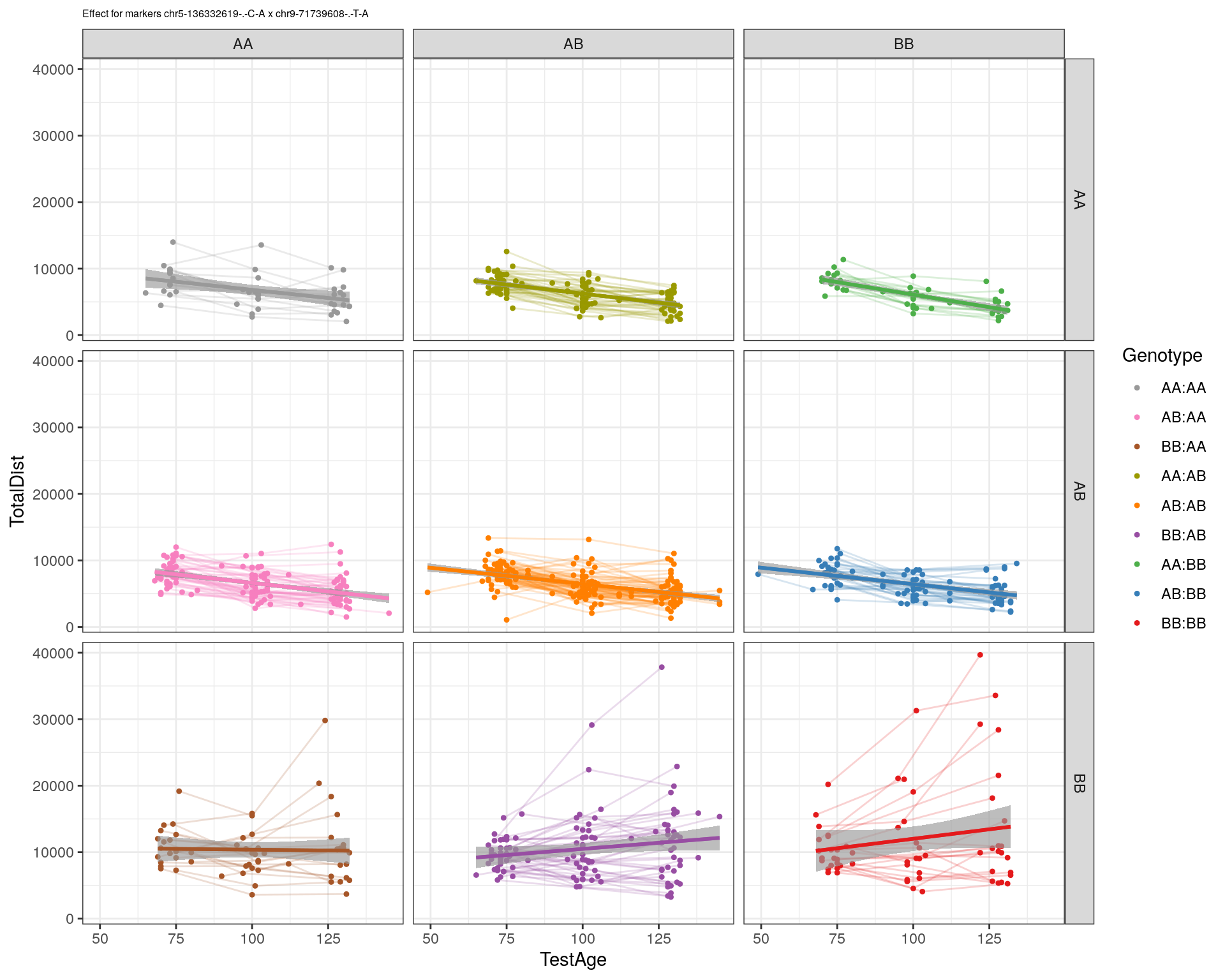
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr9-80465702-.-T-A"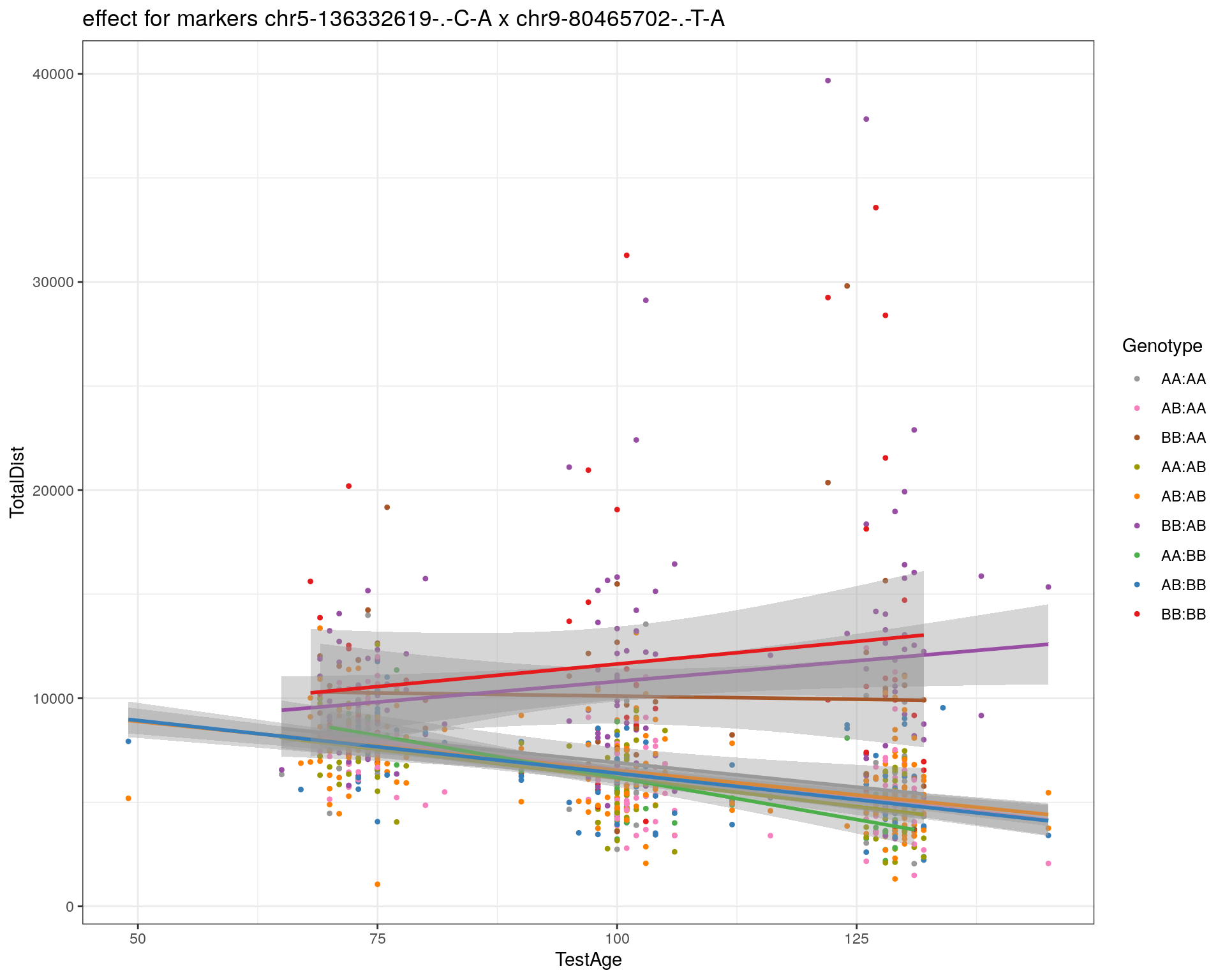
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
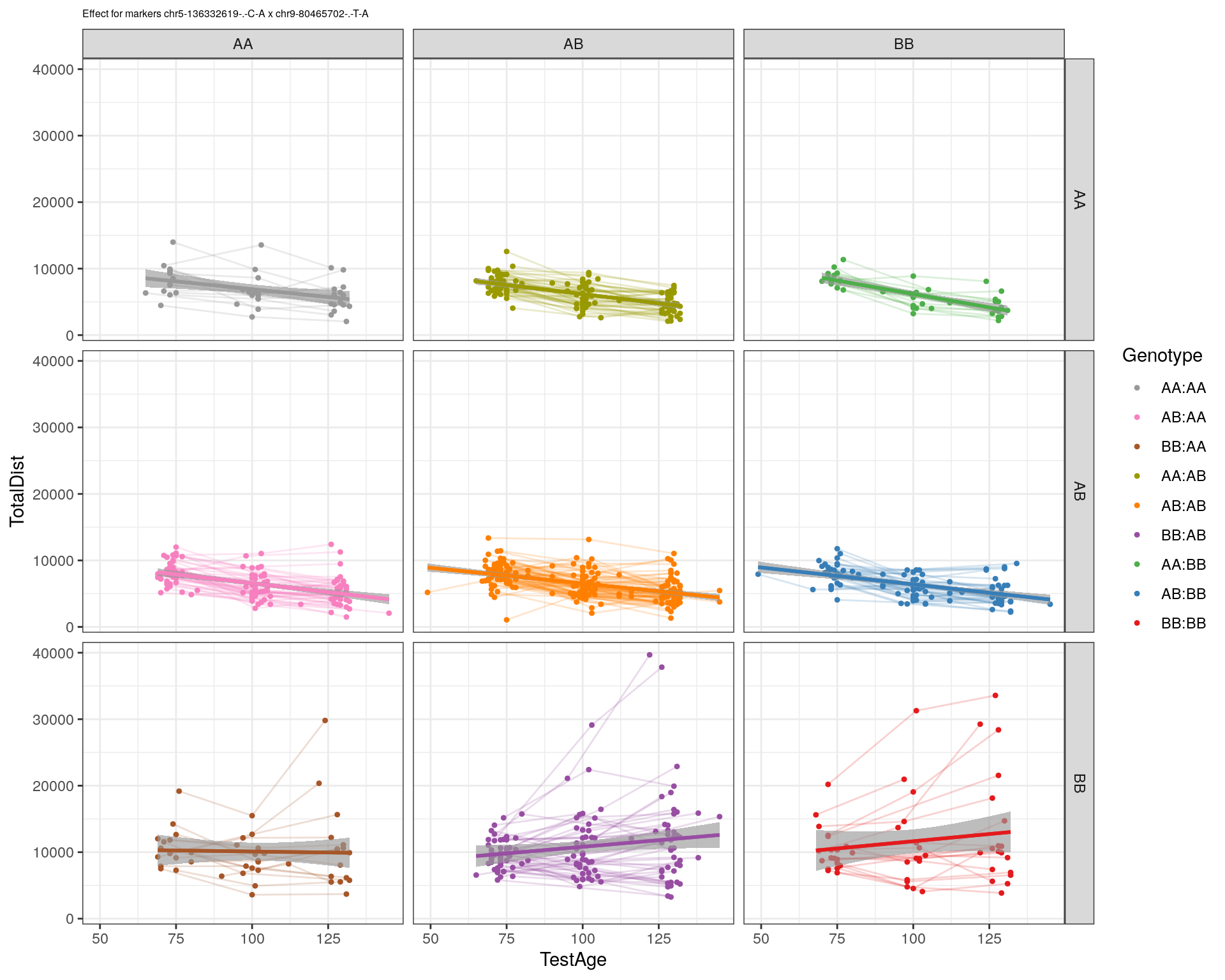
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr10-24134777-.-A-T"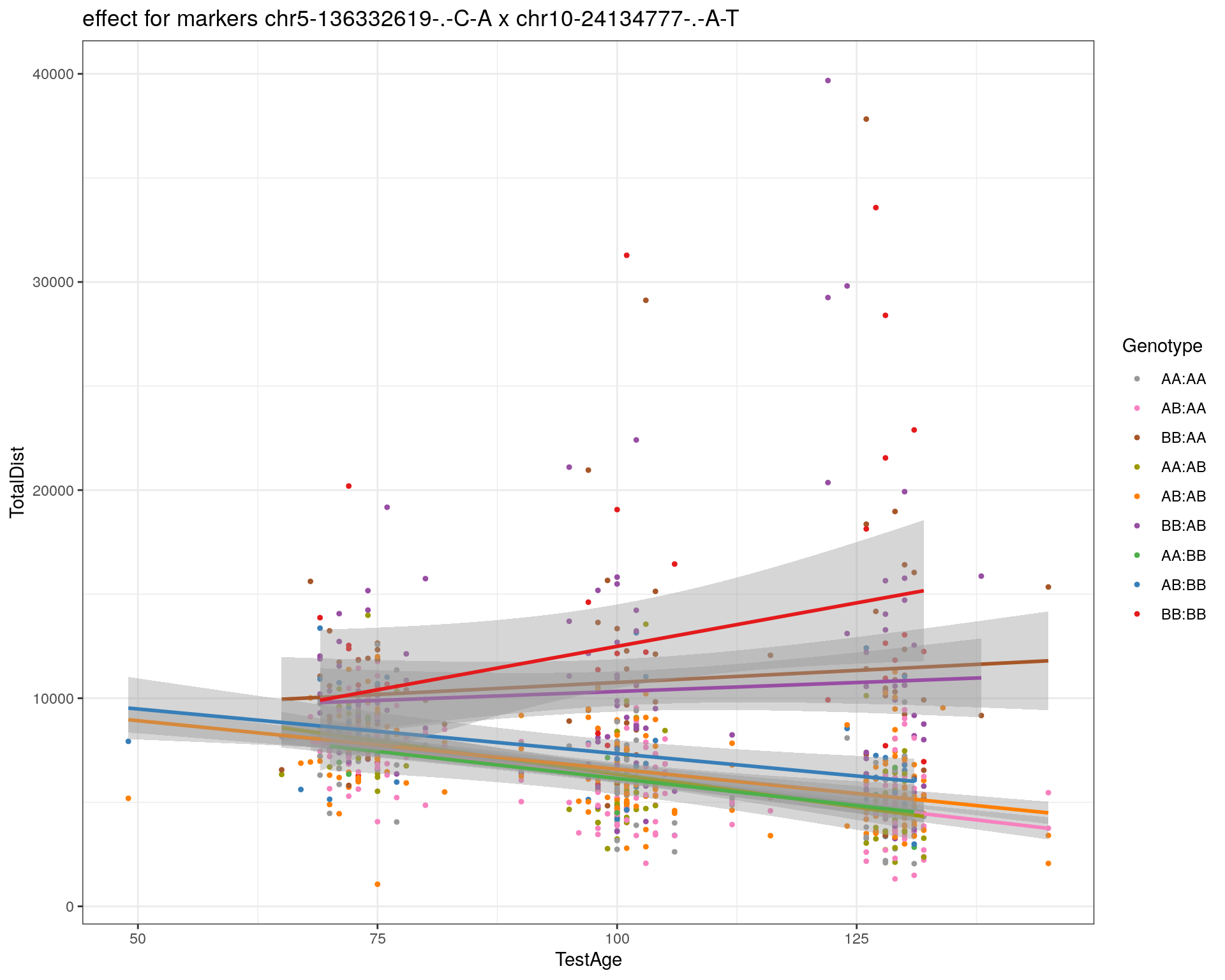
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
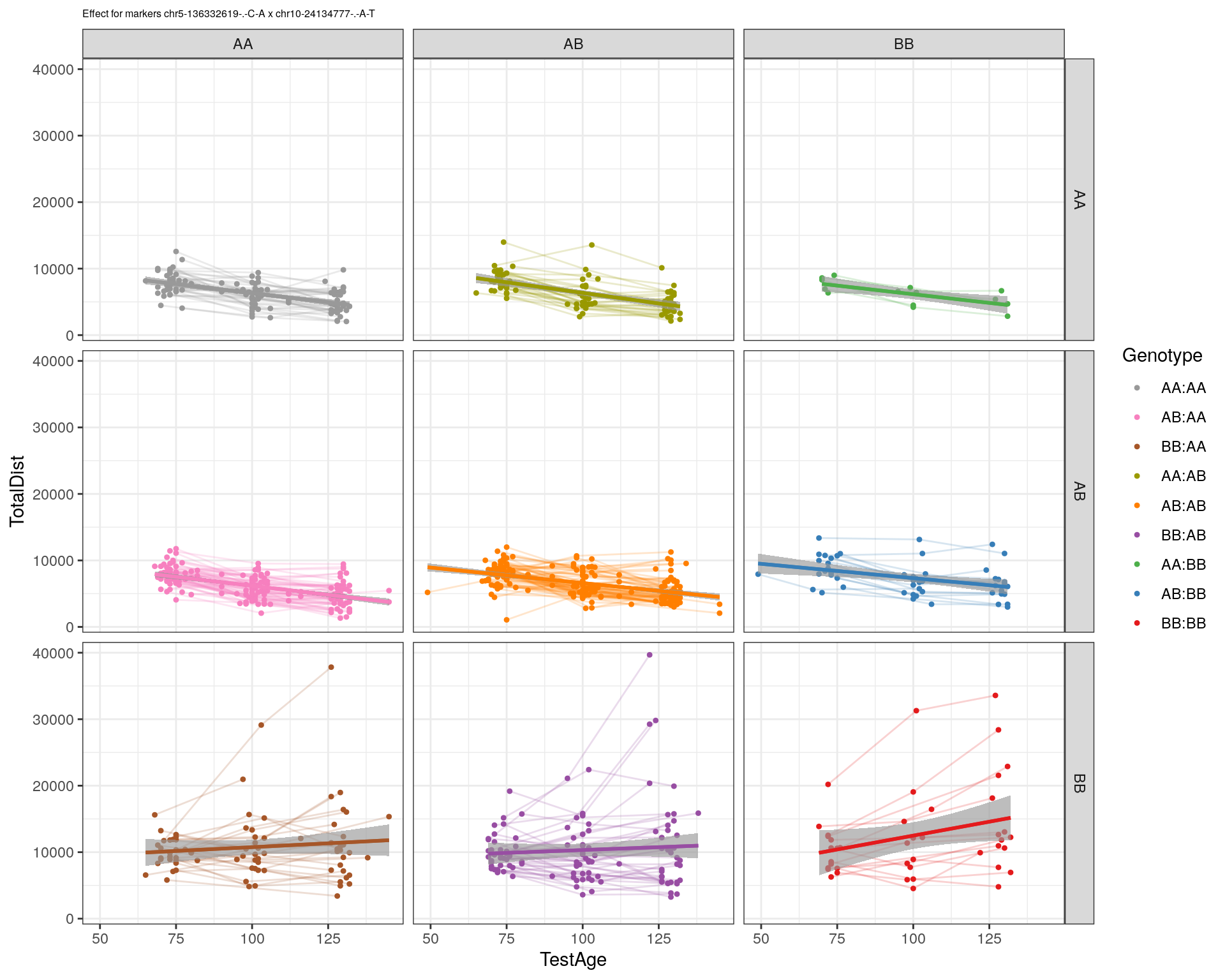
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
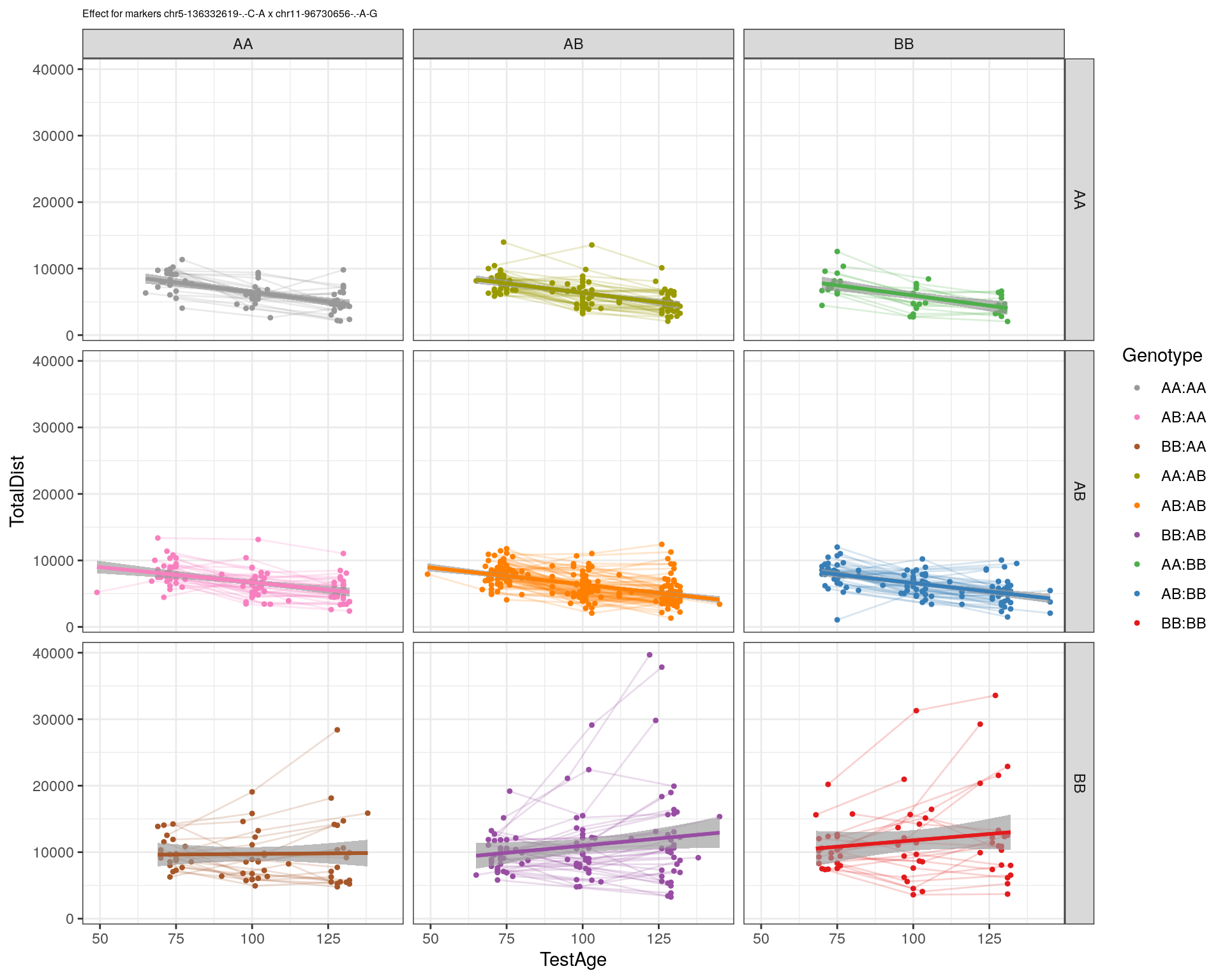
[1] "chr11-96730656-.-A-G"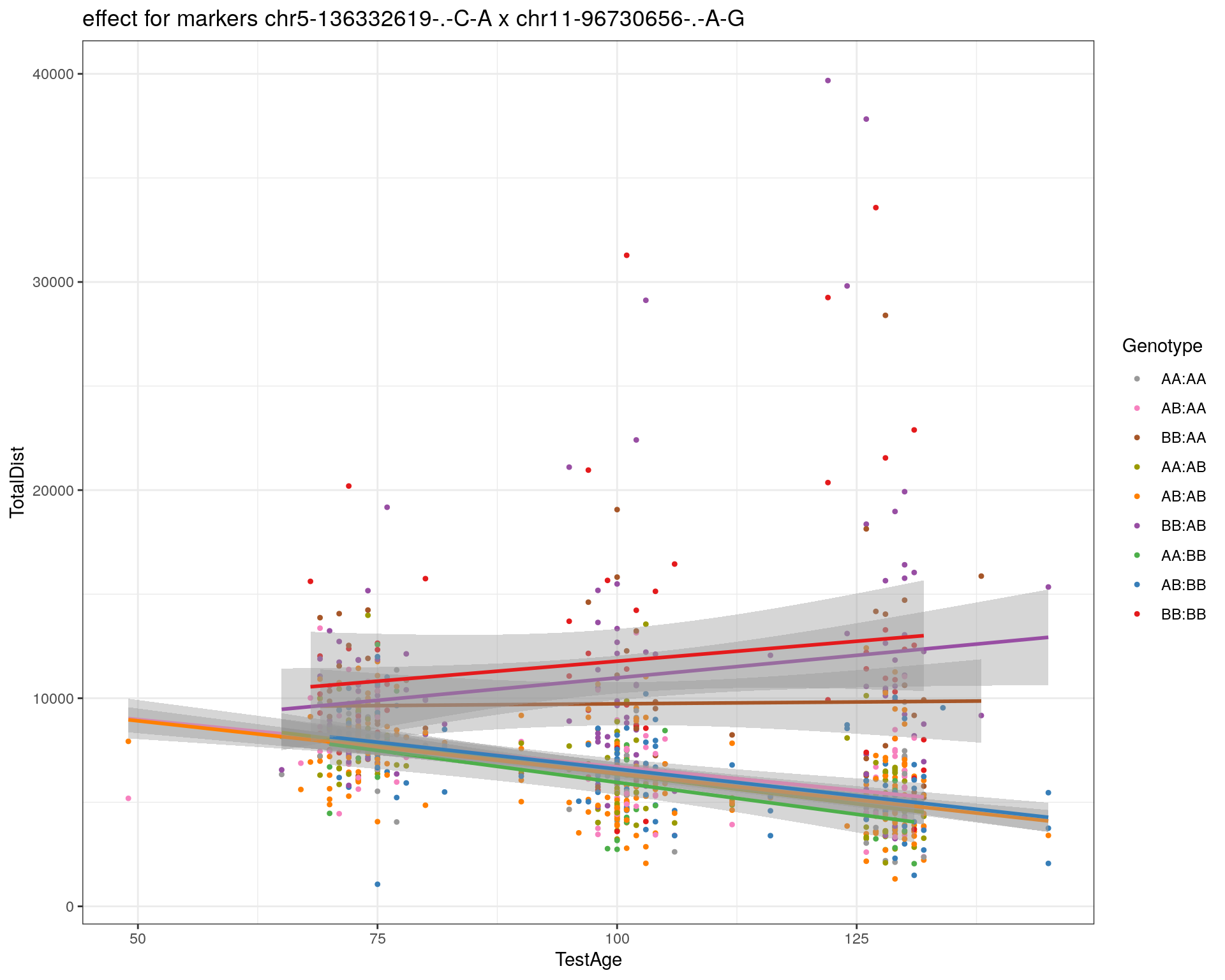
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
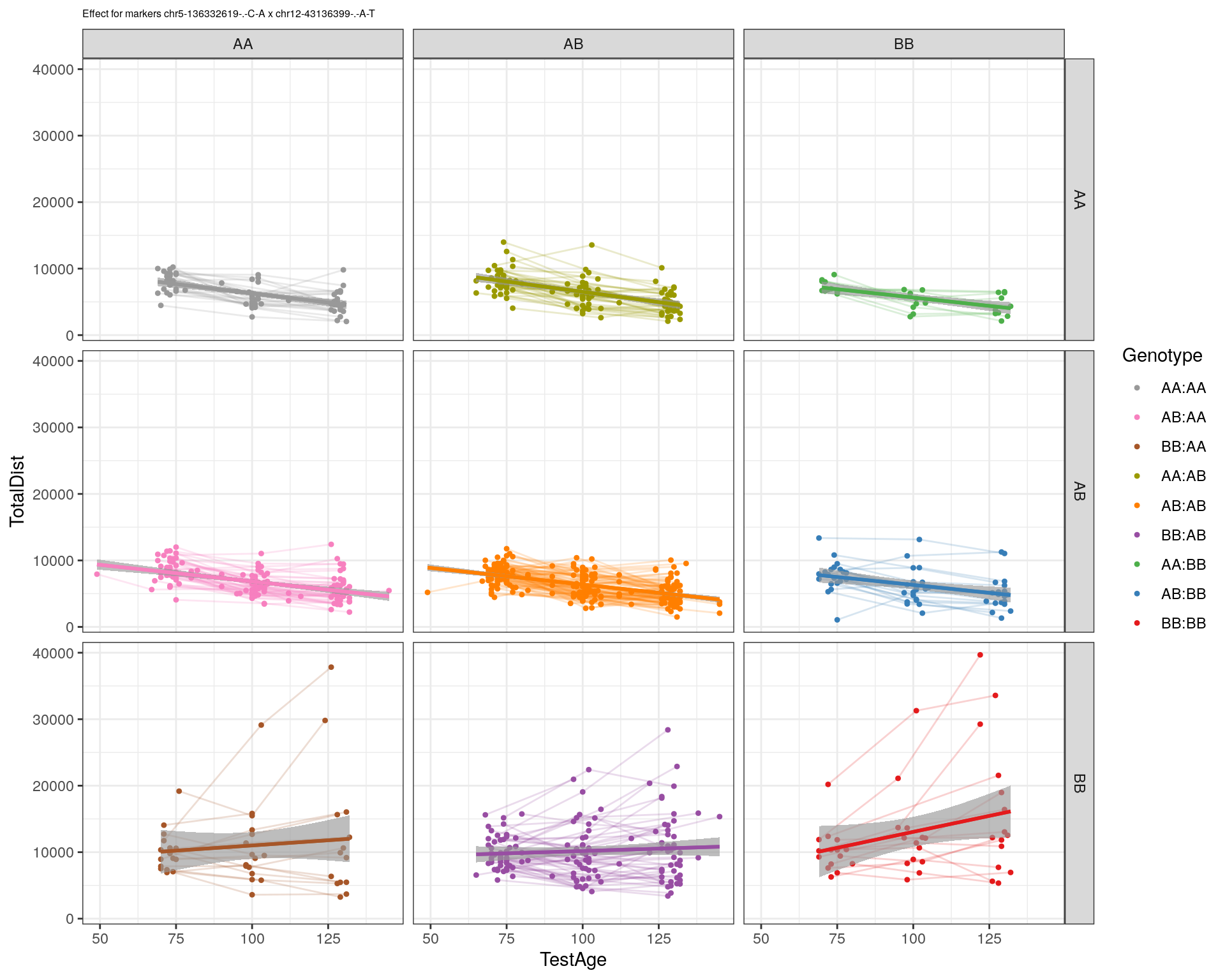
[1] "chr12-43136399-.-A-T"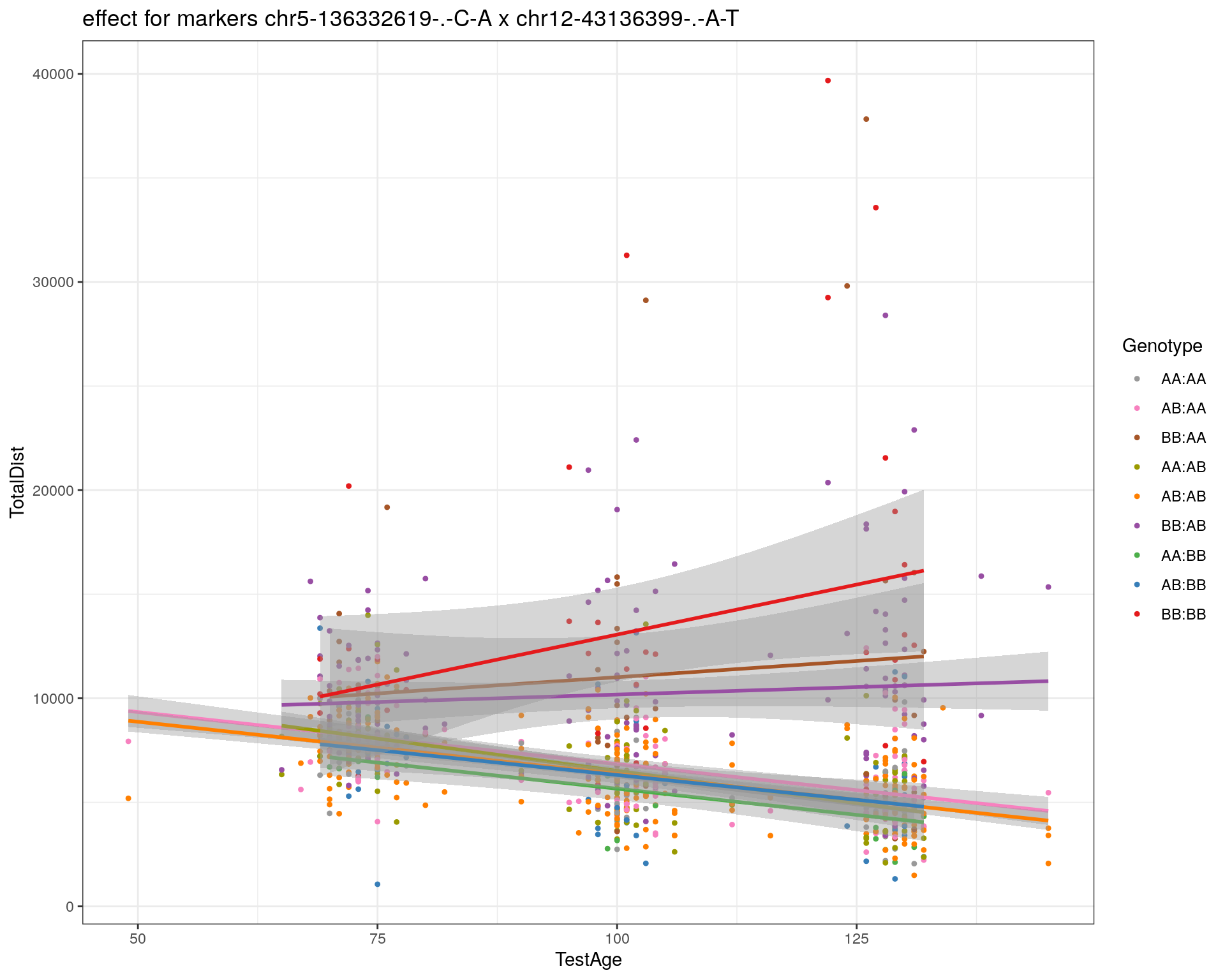
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr12-69197484-.-C-A"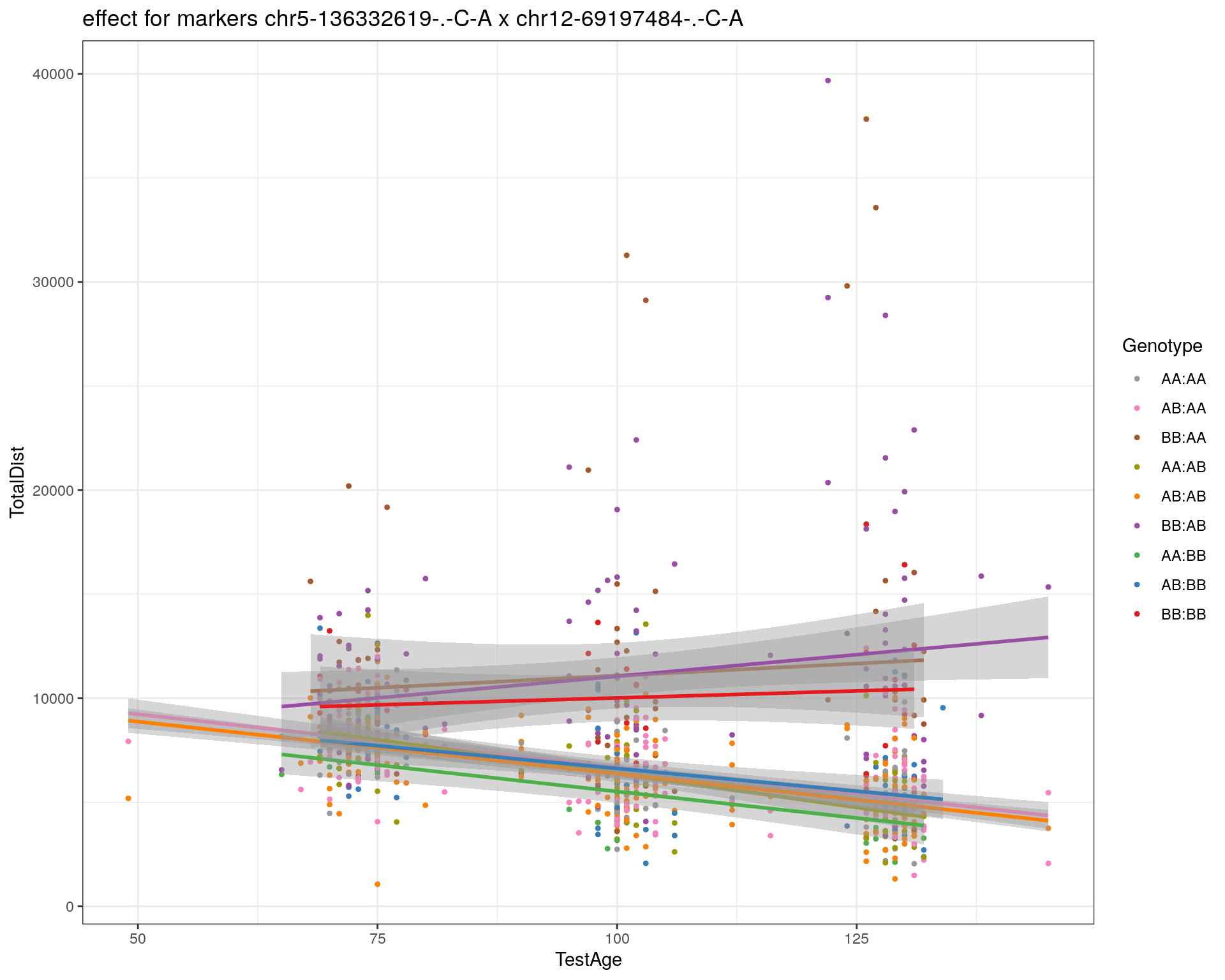
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
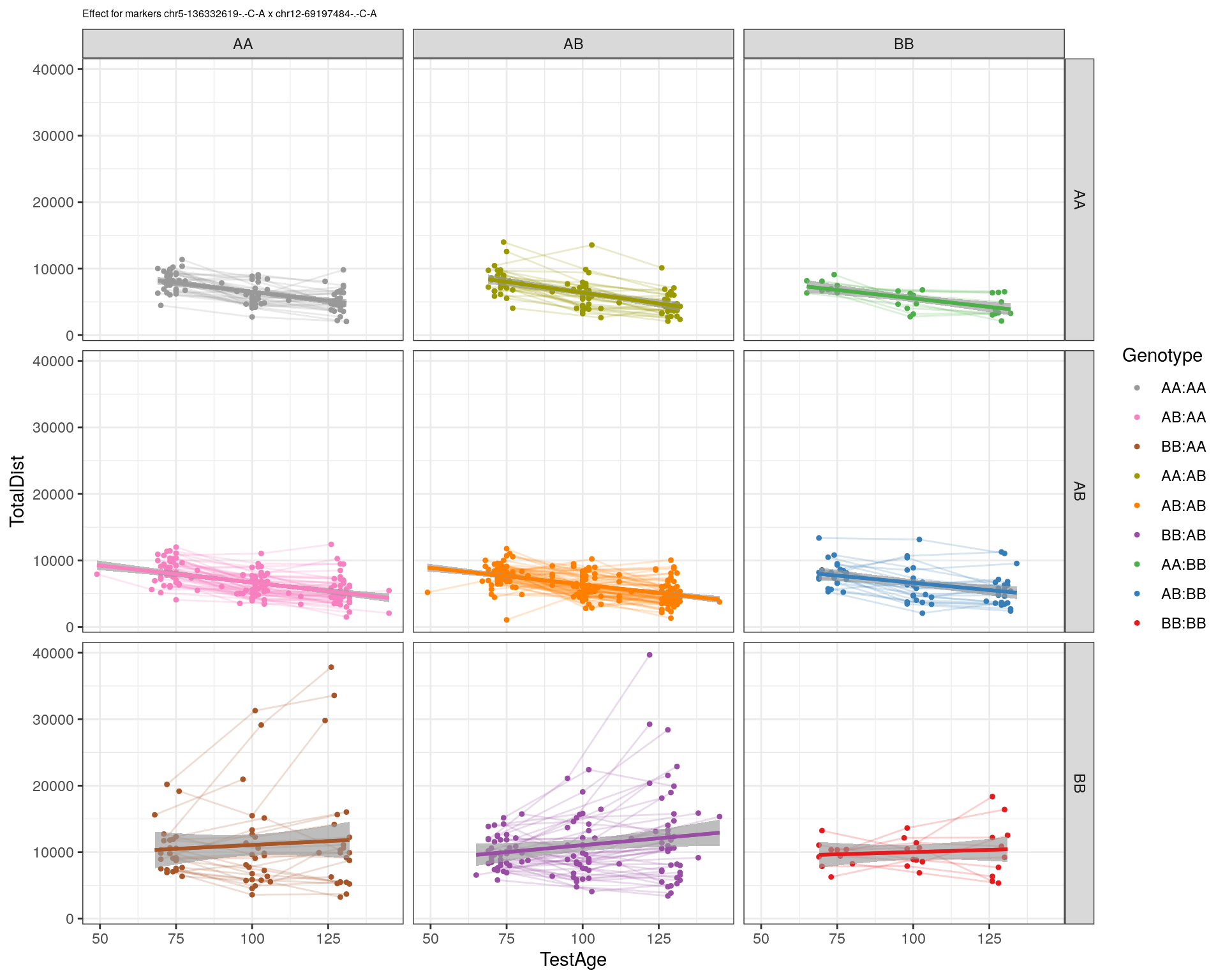
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr12-86851130-.-T-A"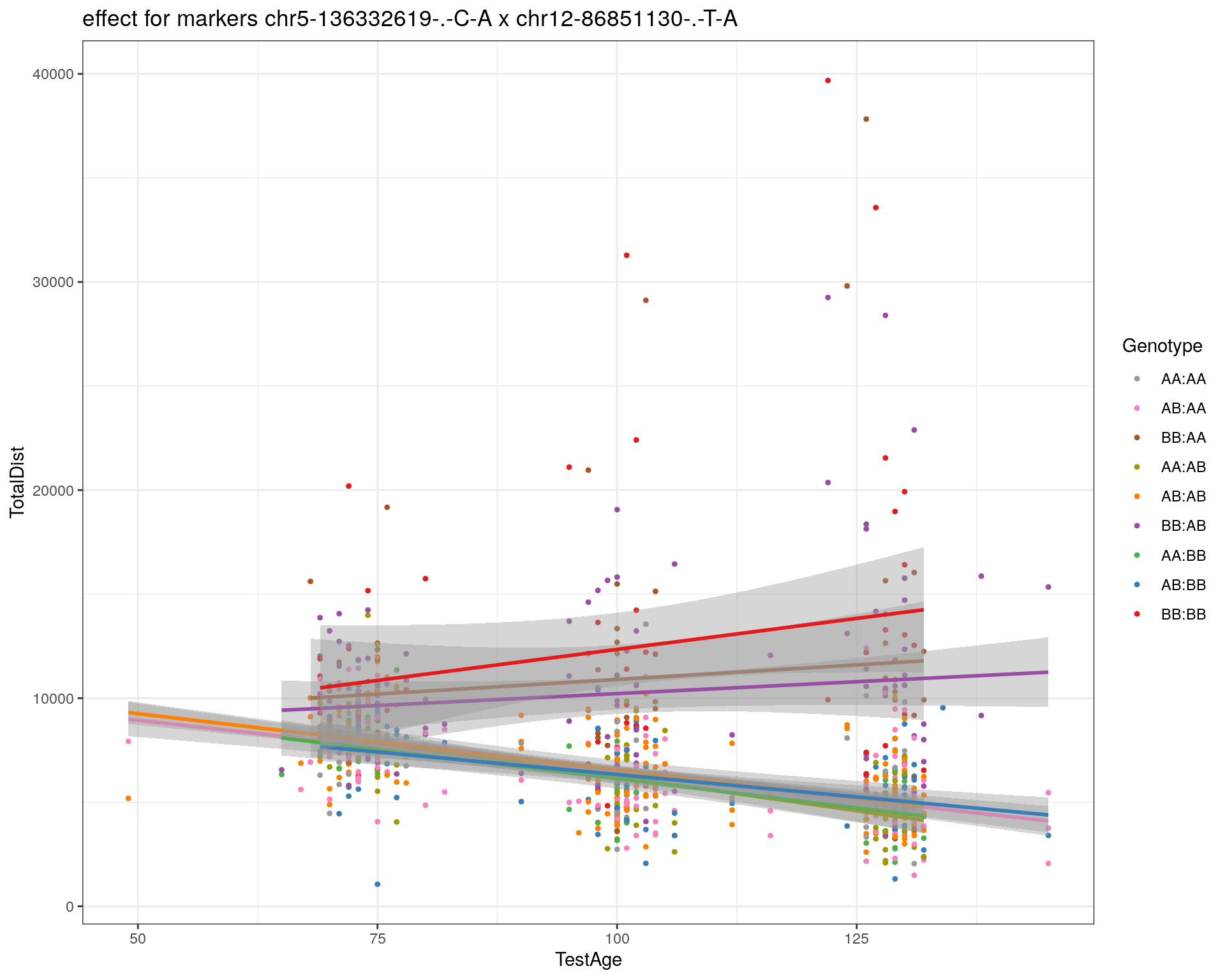
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
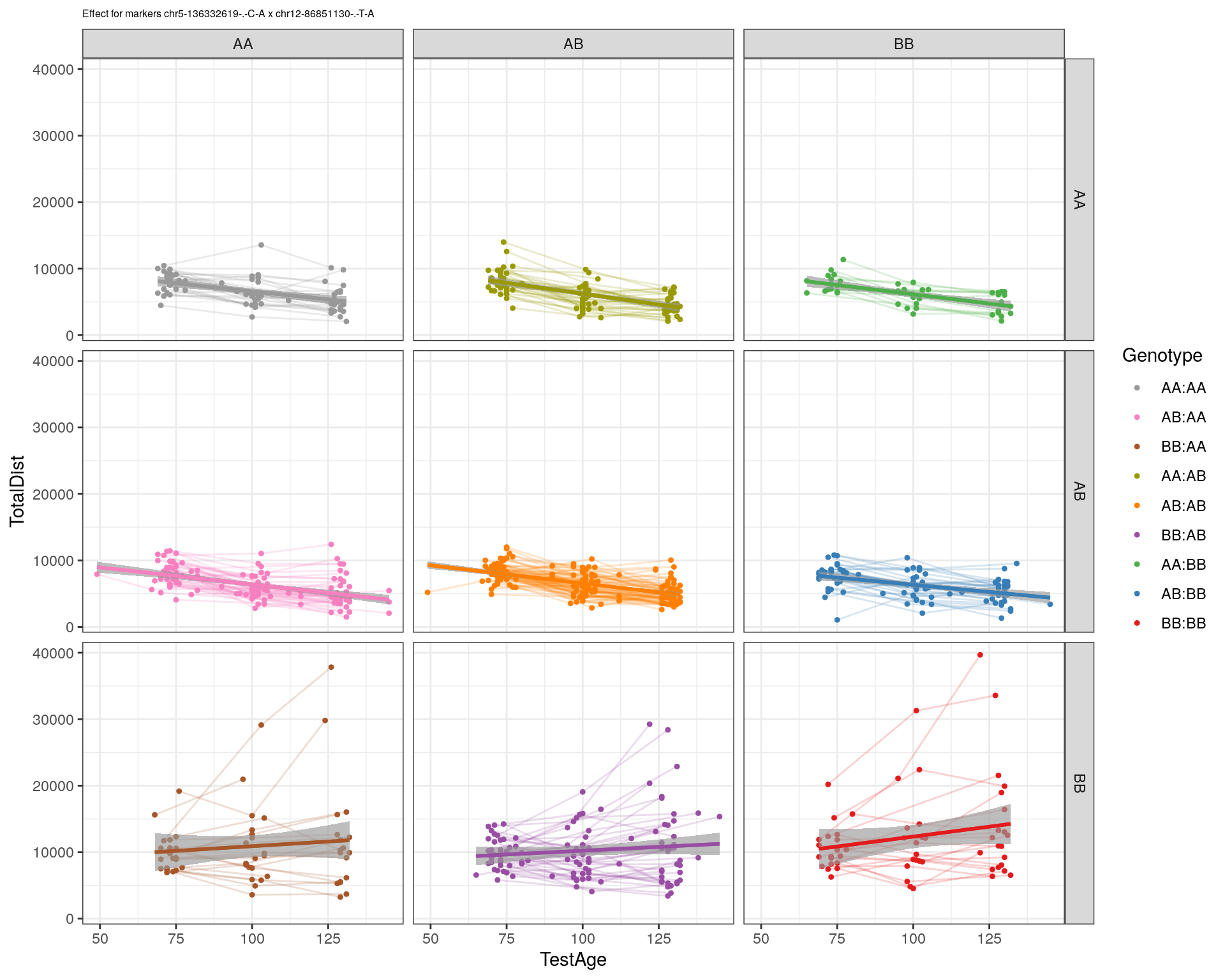
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
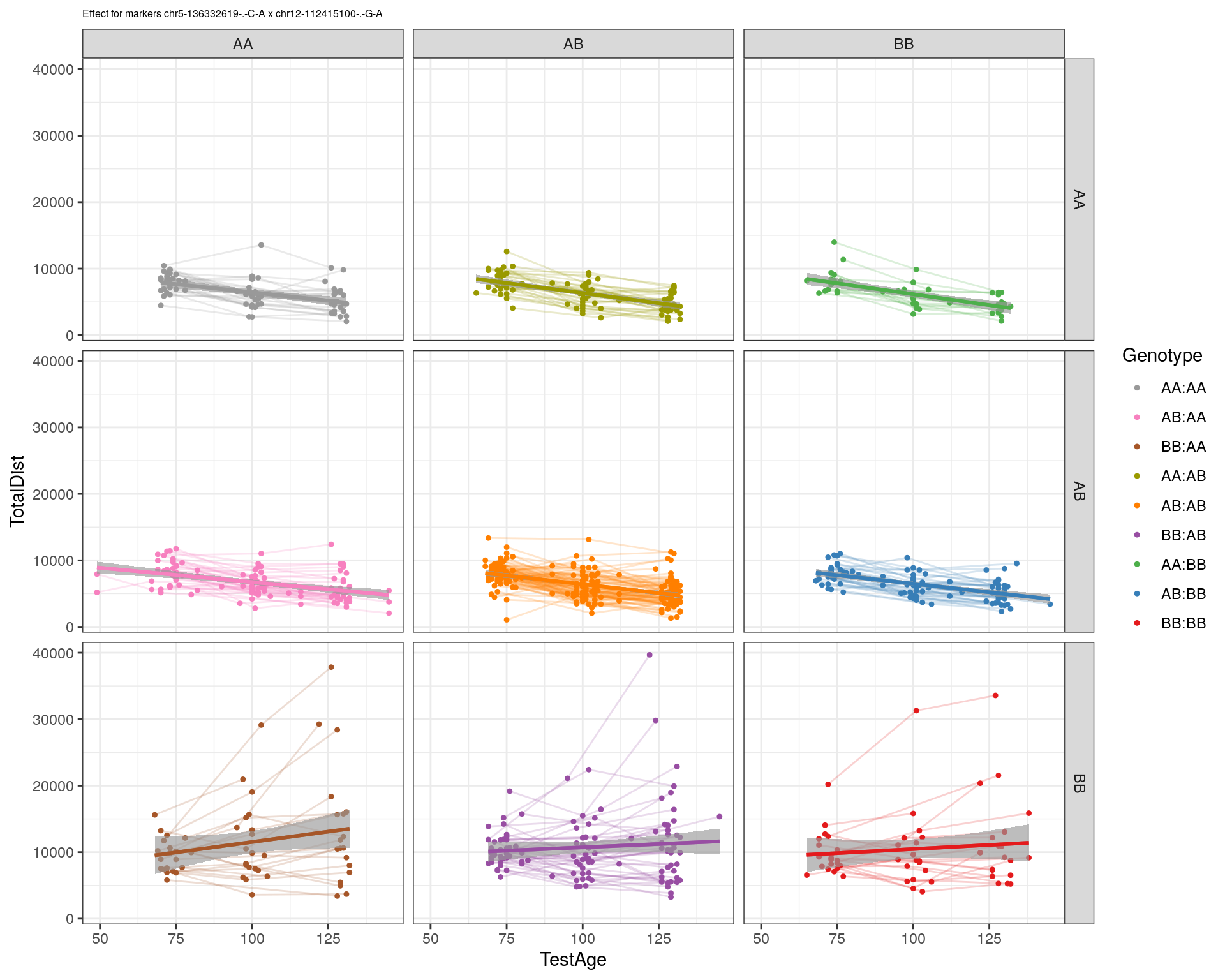
[1] "chr12-112415100-.-G-A"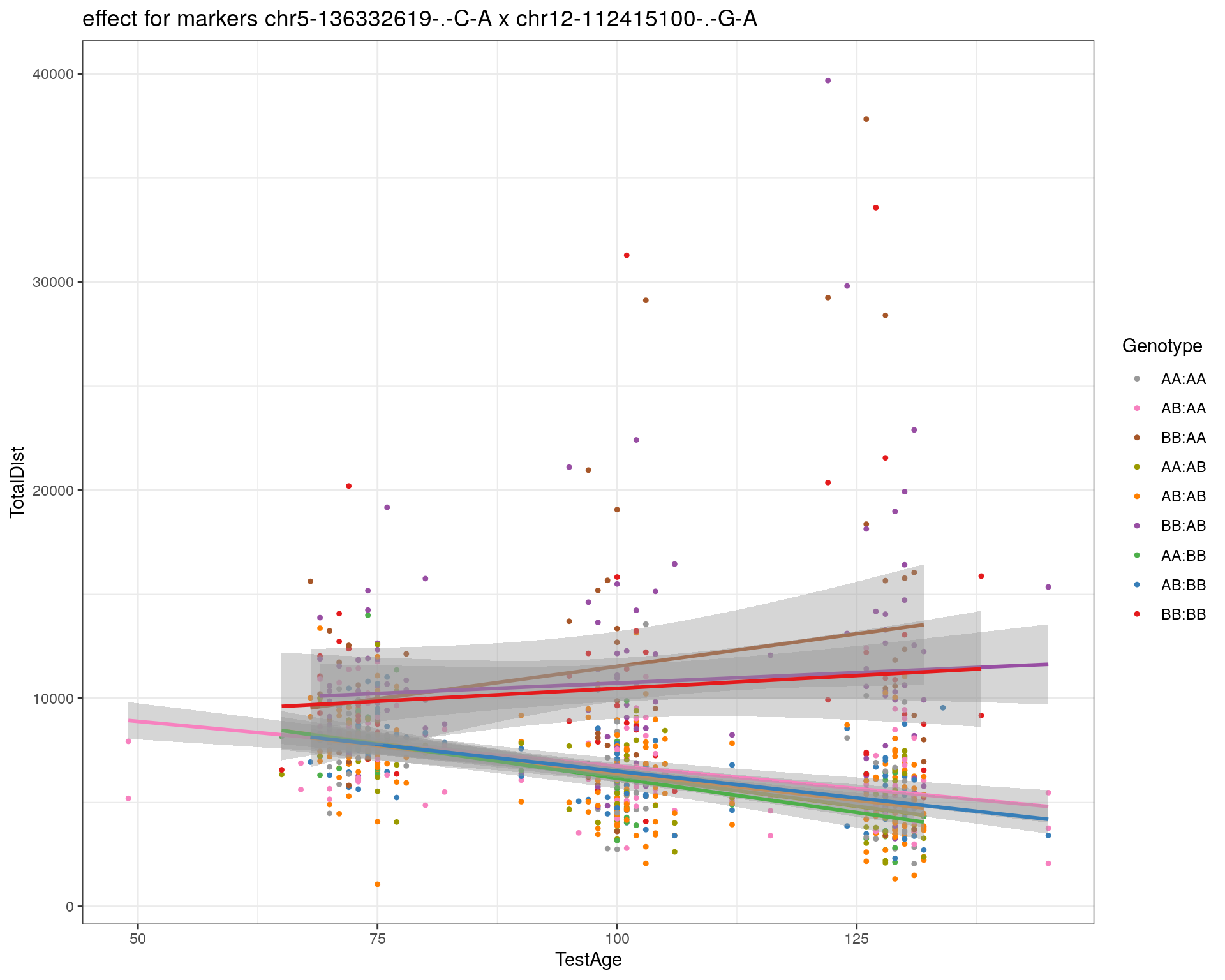
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
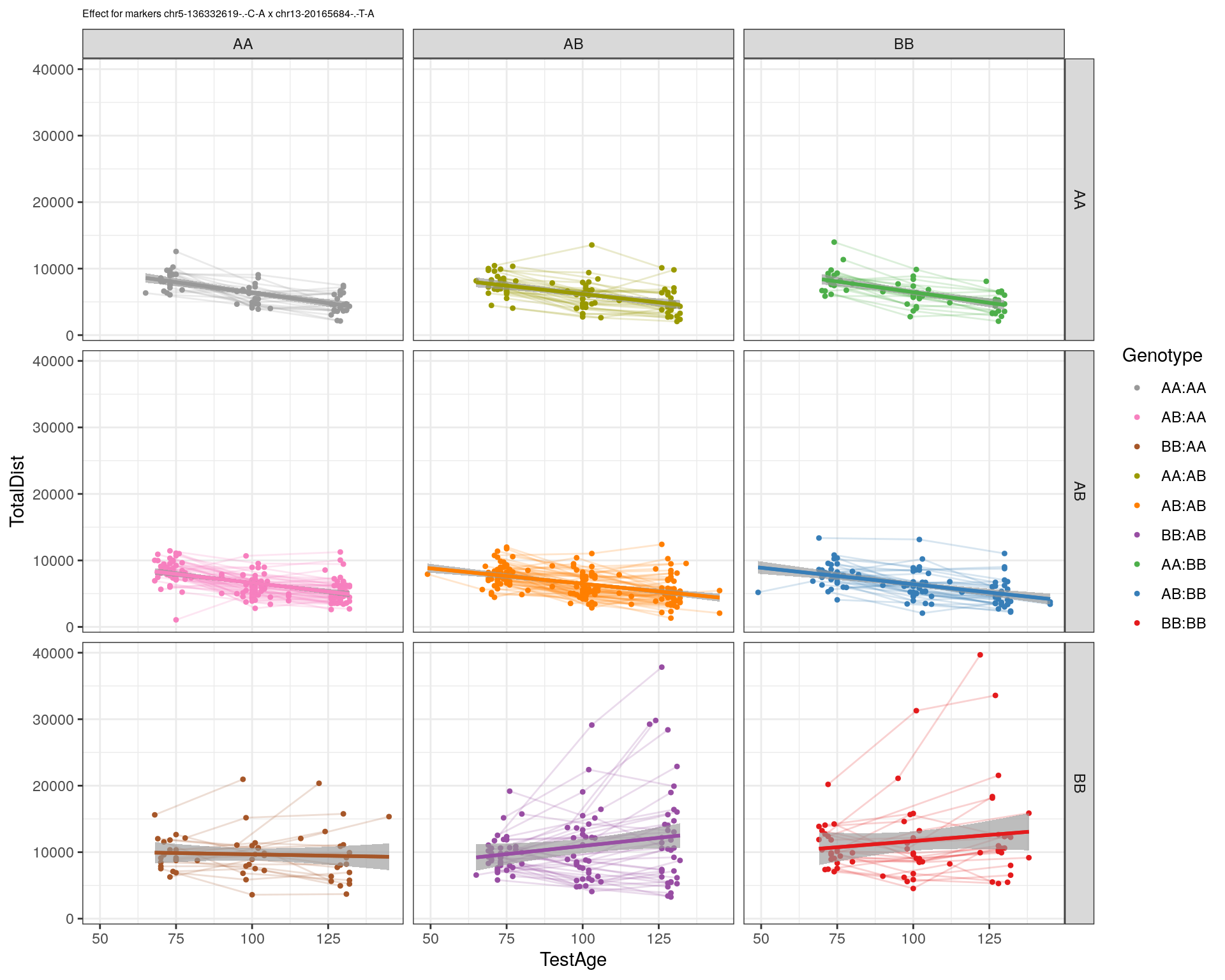
[1] "chr13-20165684-.-T-A"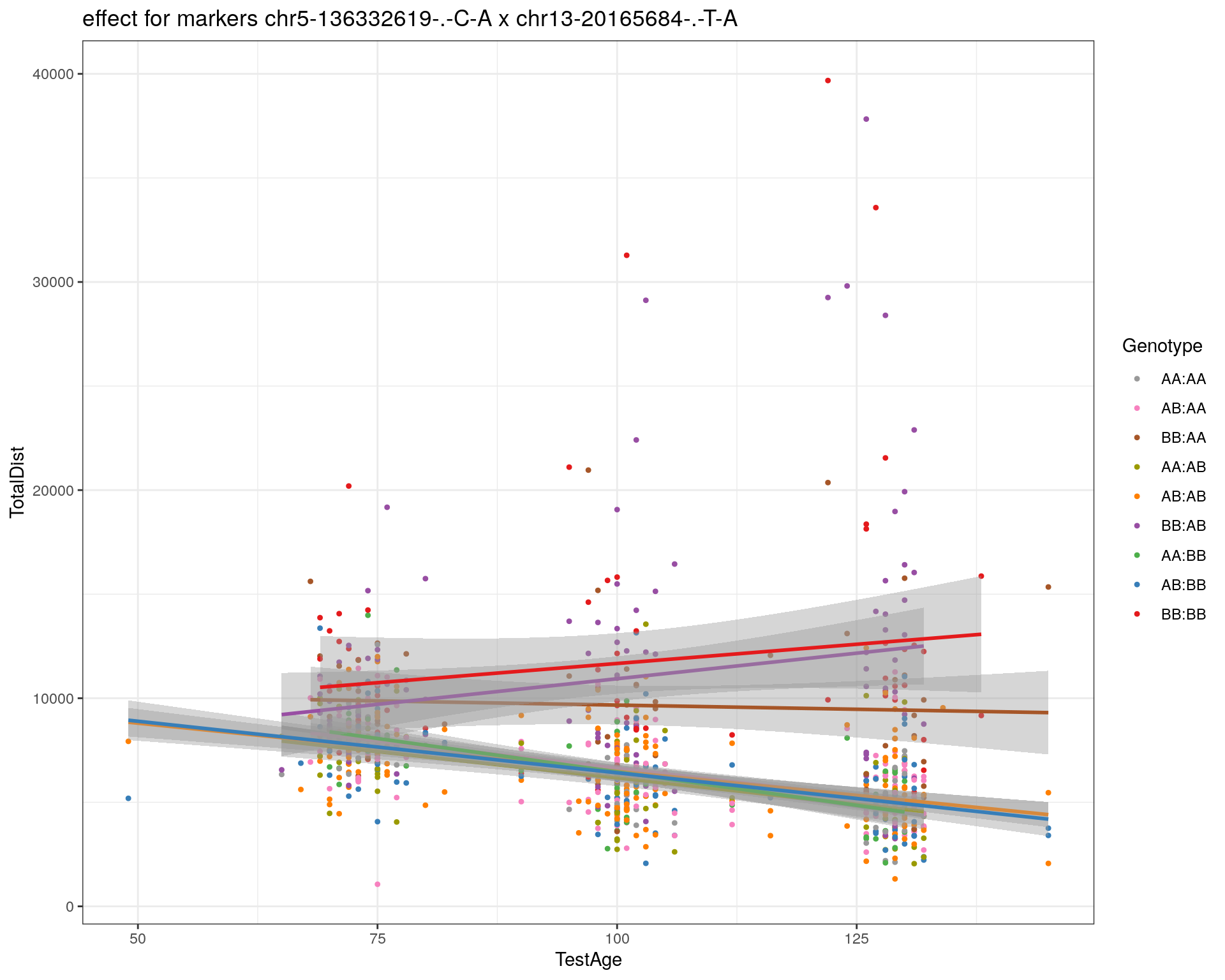
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
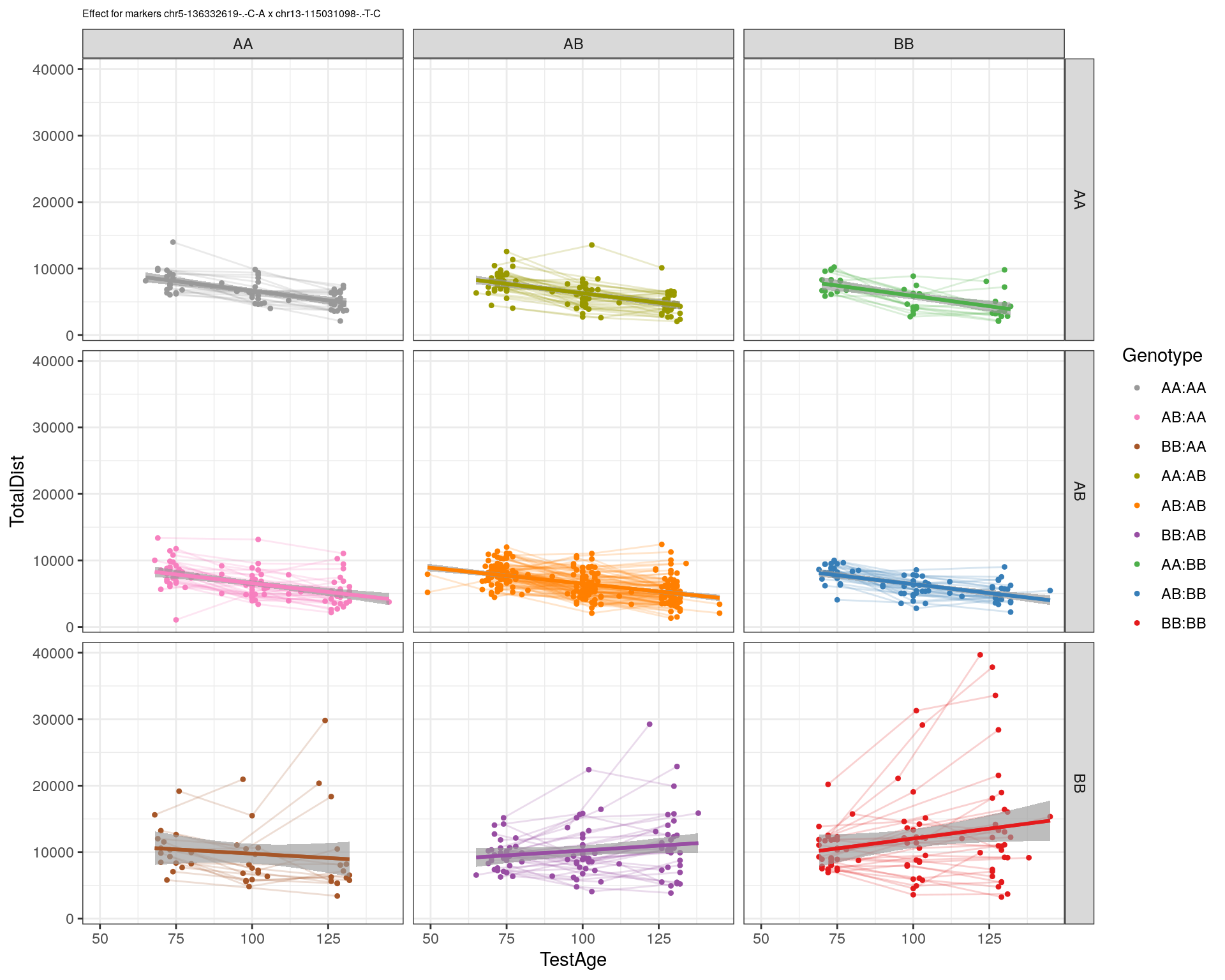
[1] "chr13-115031098-.-T-C"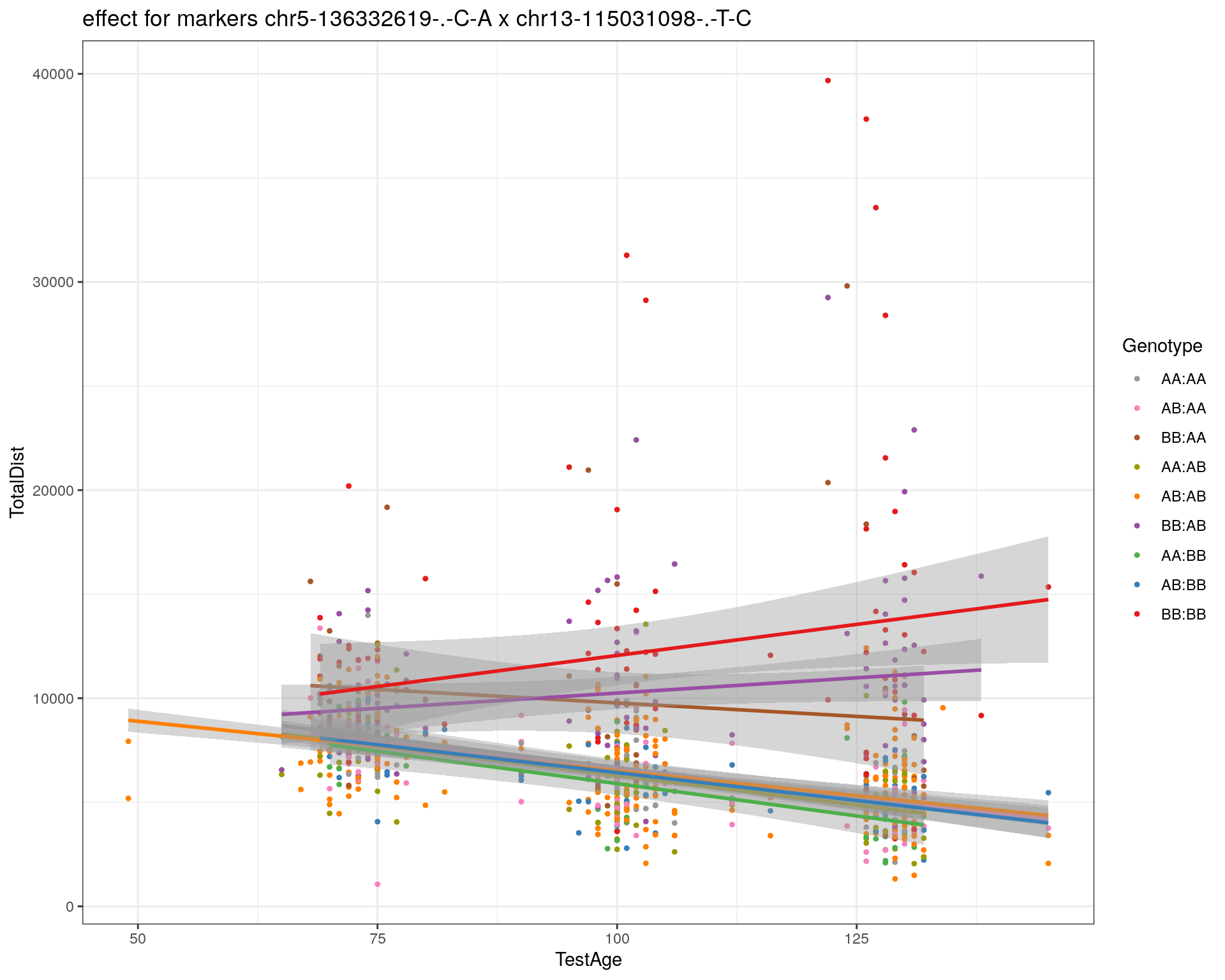
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
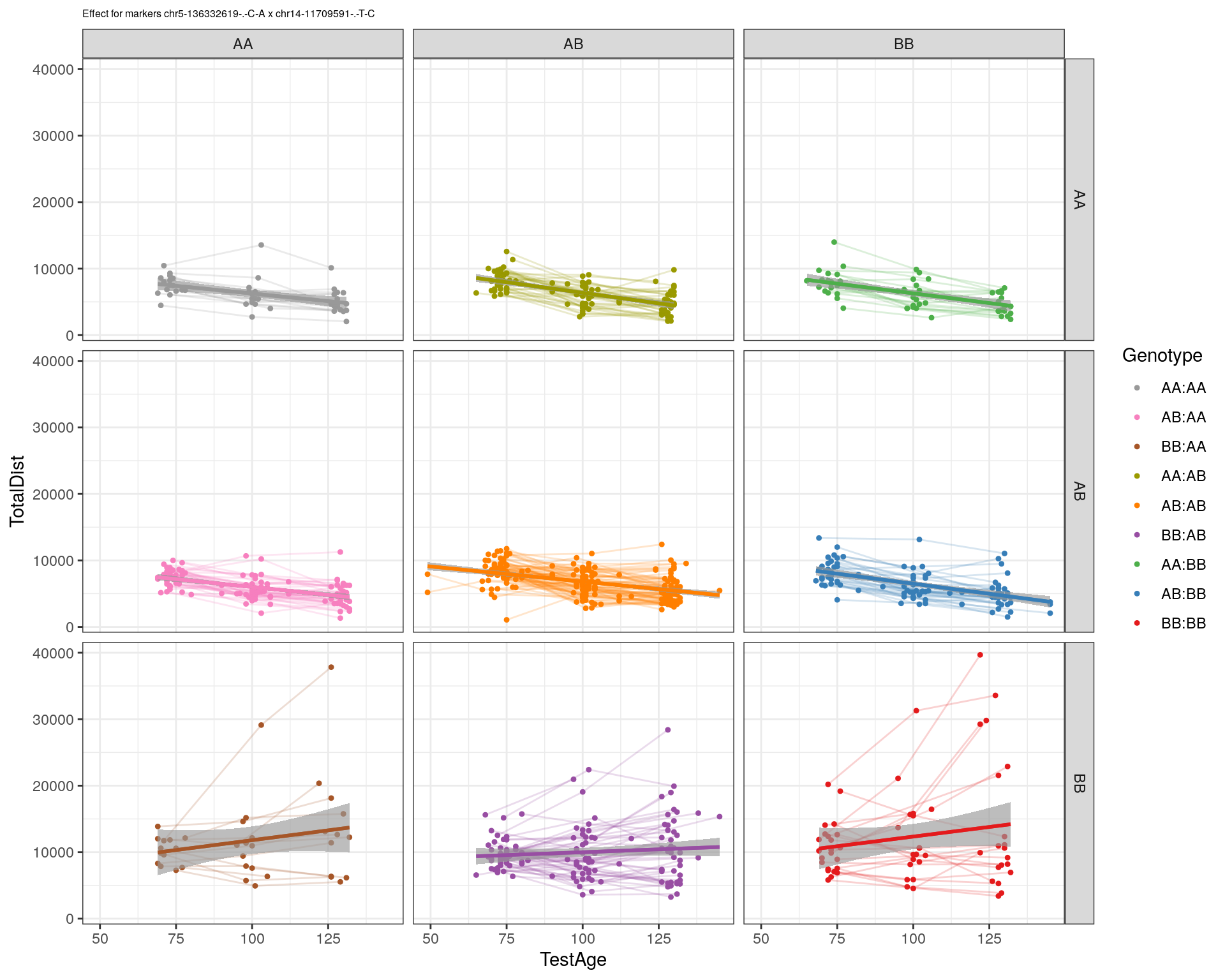
[1] "chr14-11709591-.-T-C"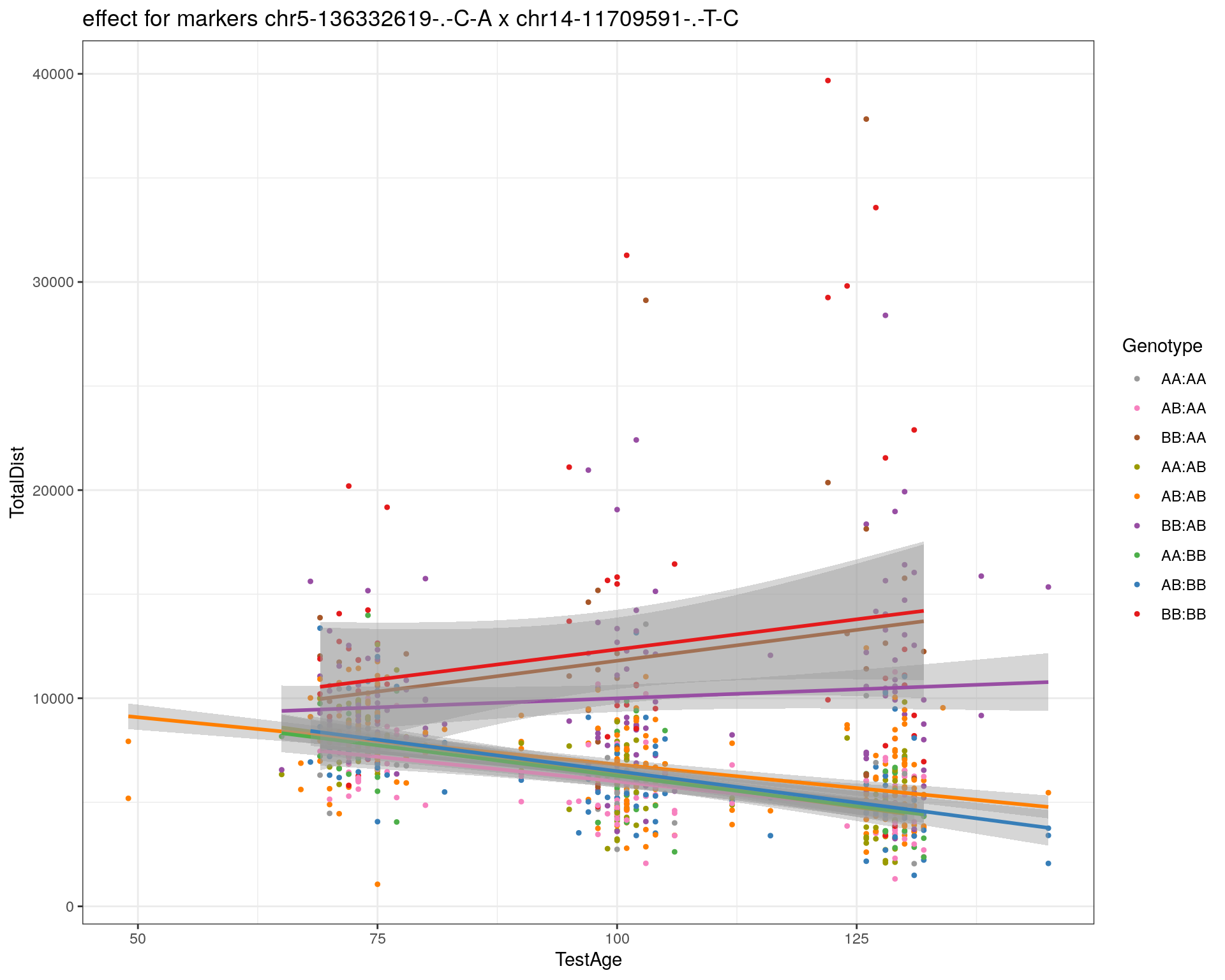
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr14-50642778-.-G-A"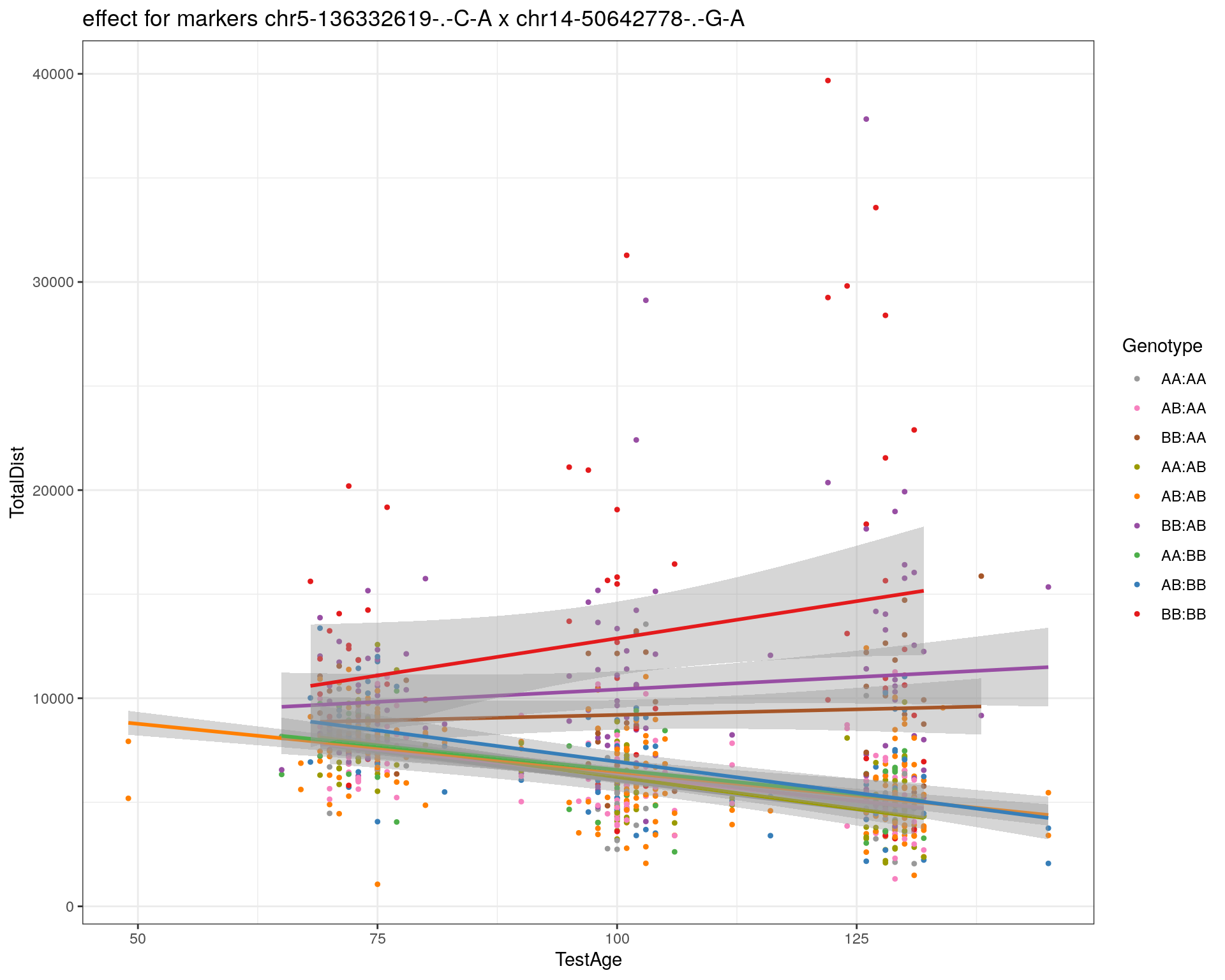
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
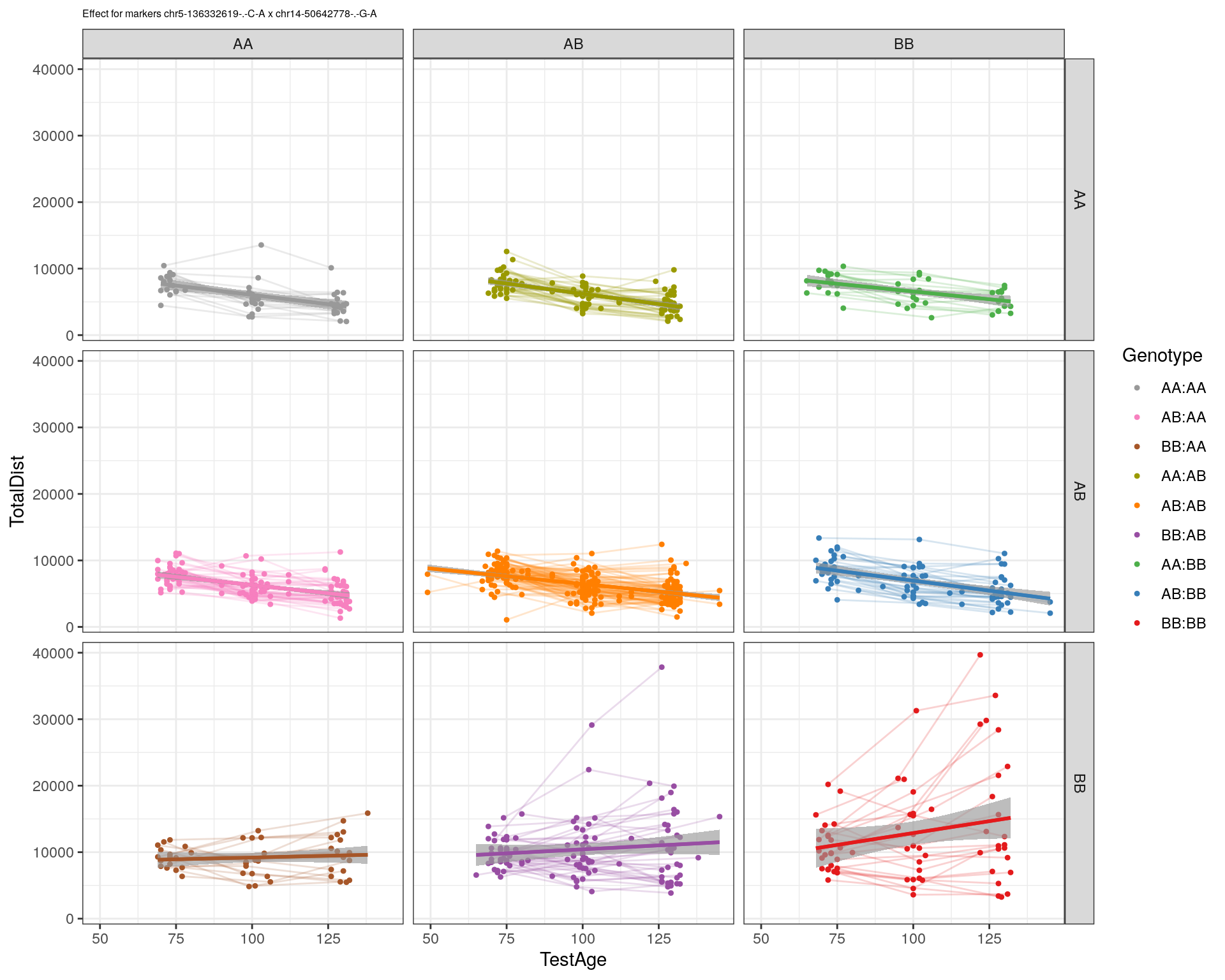
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr14-62407765-.-T-A"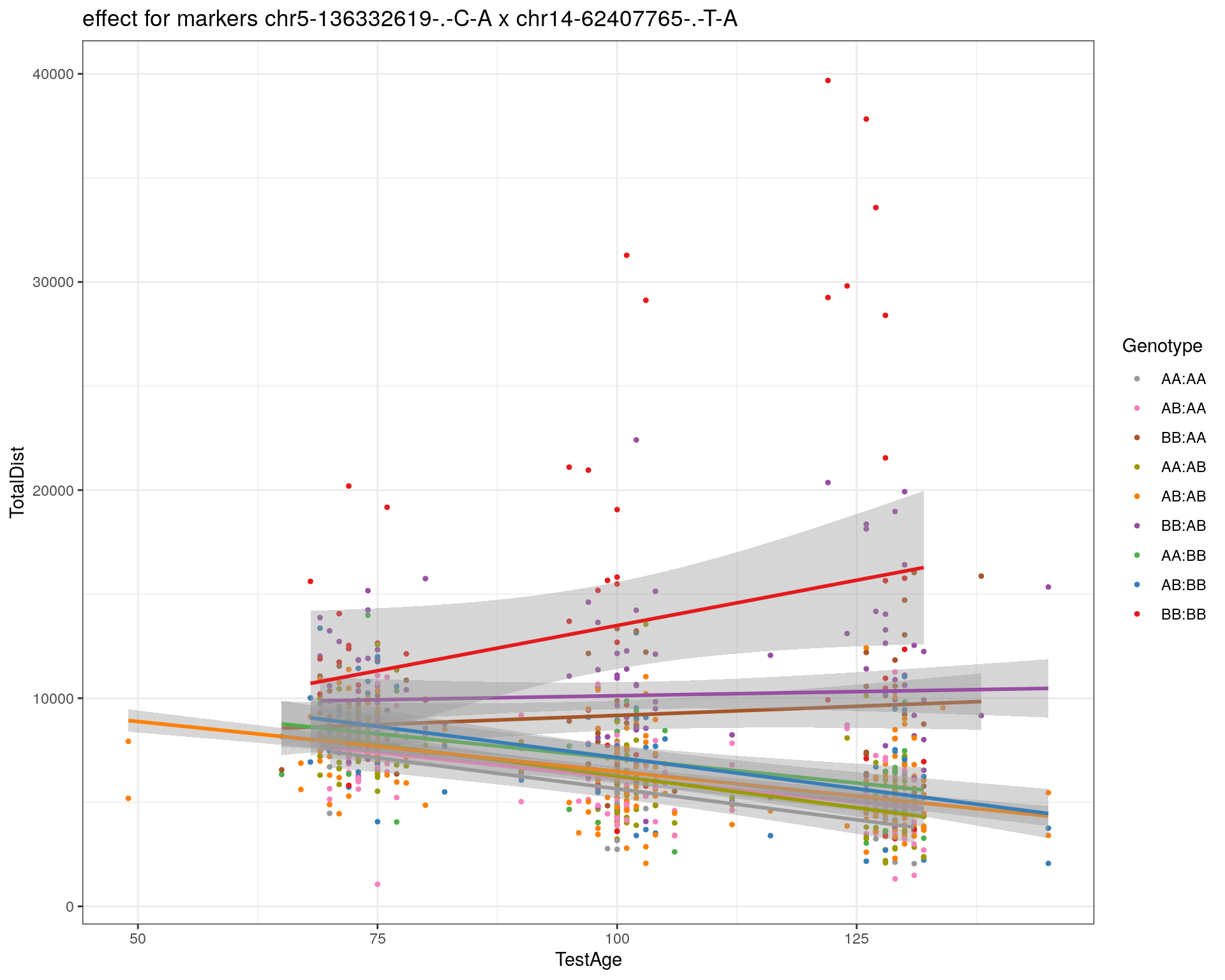
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
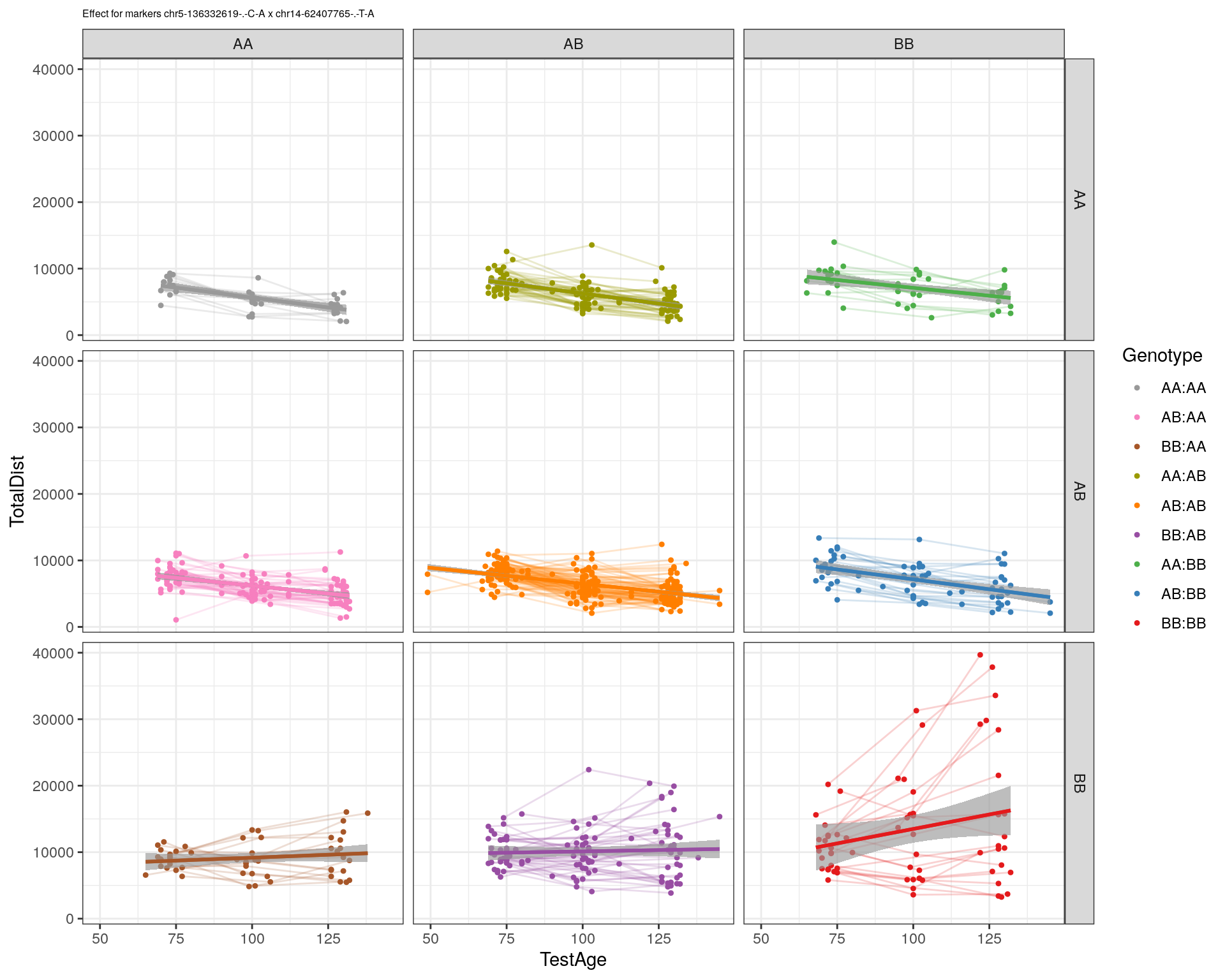
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr14-69691477-.-G-T"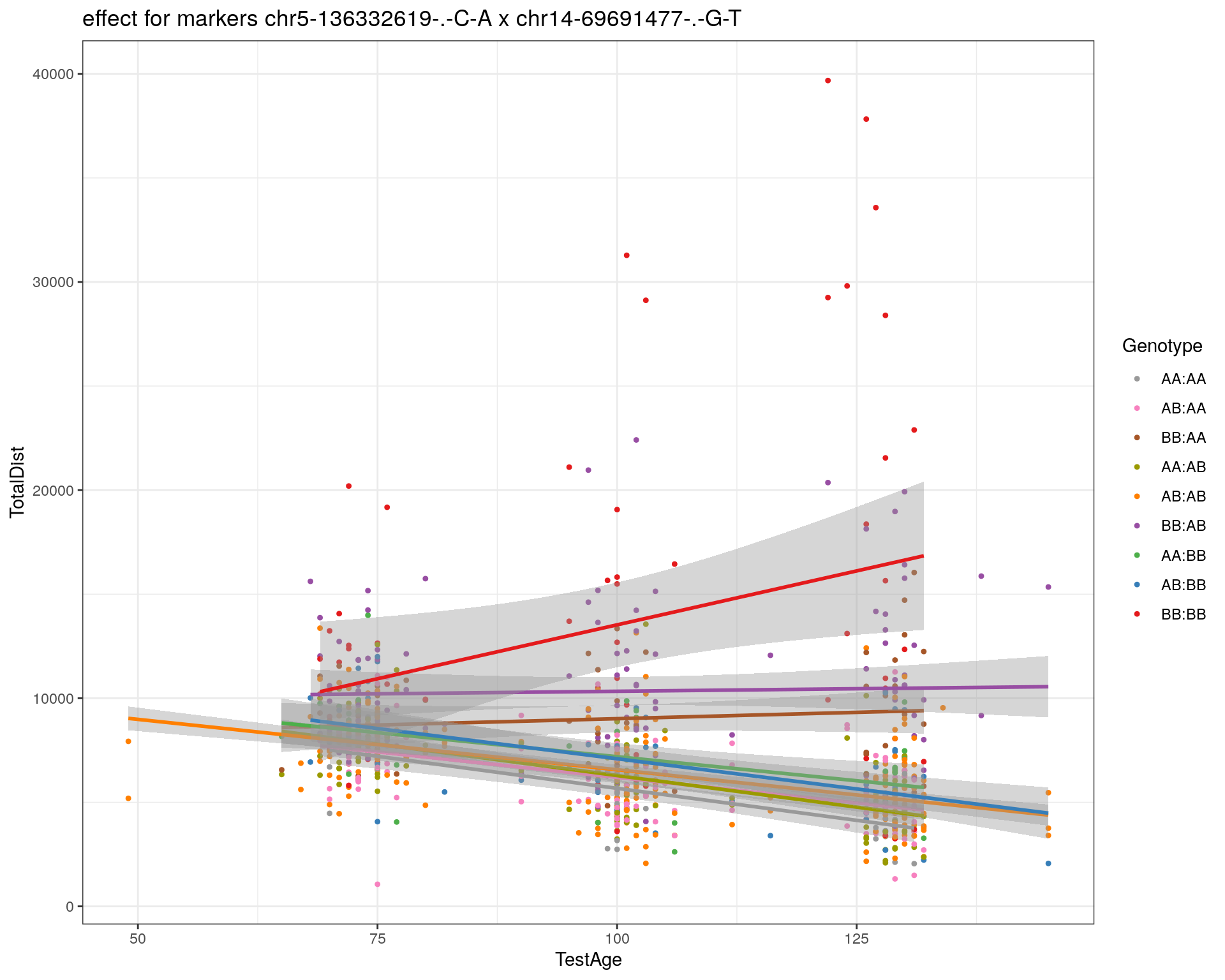
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
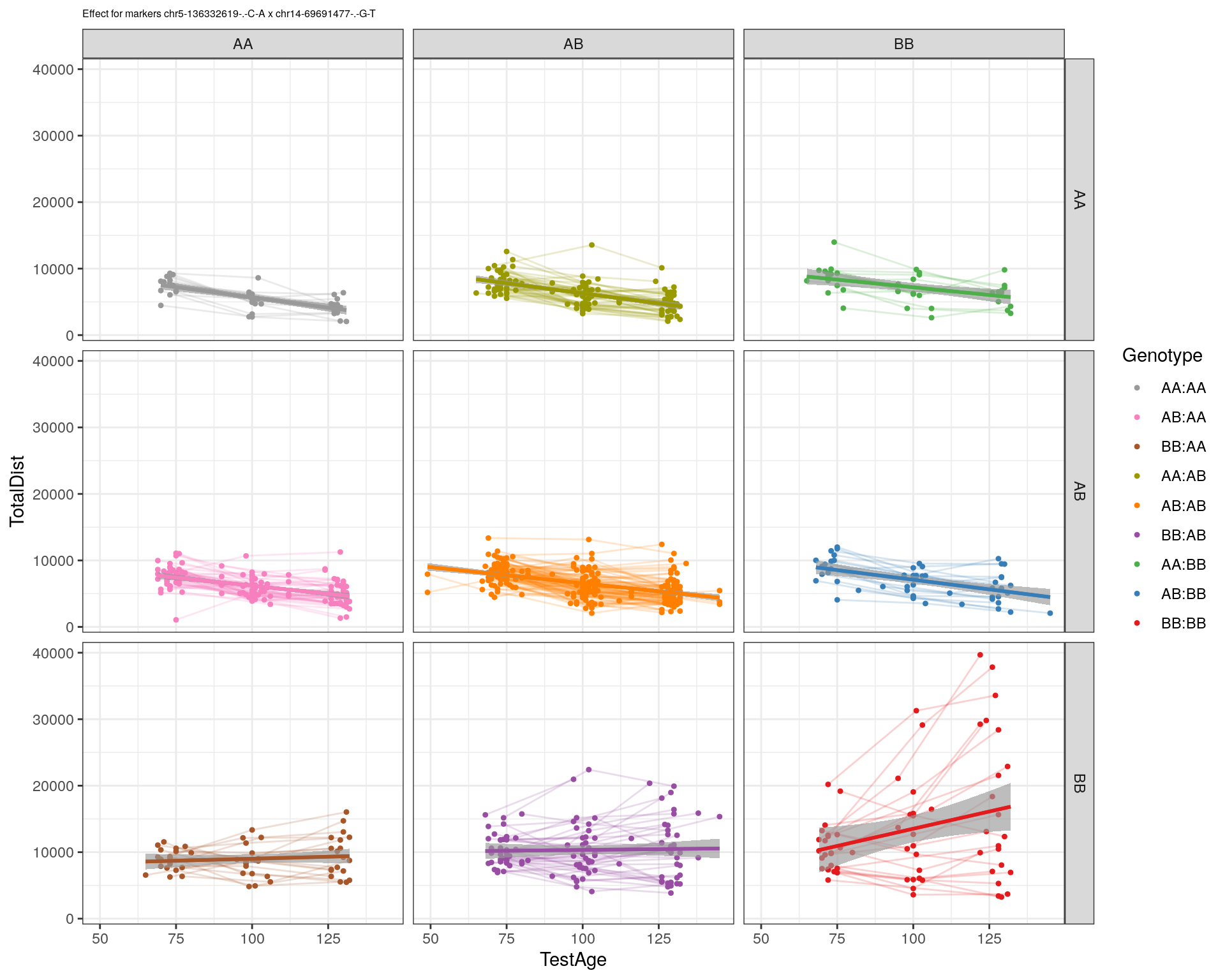
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr15-58605950-.-C-G"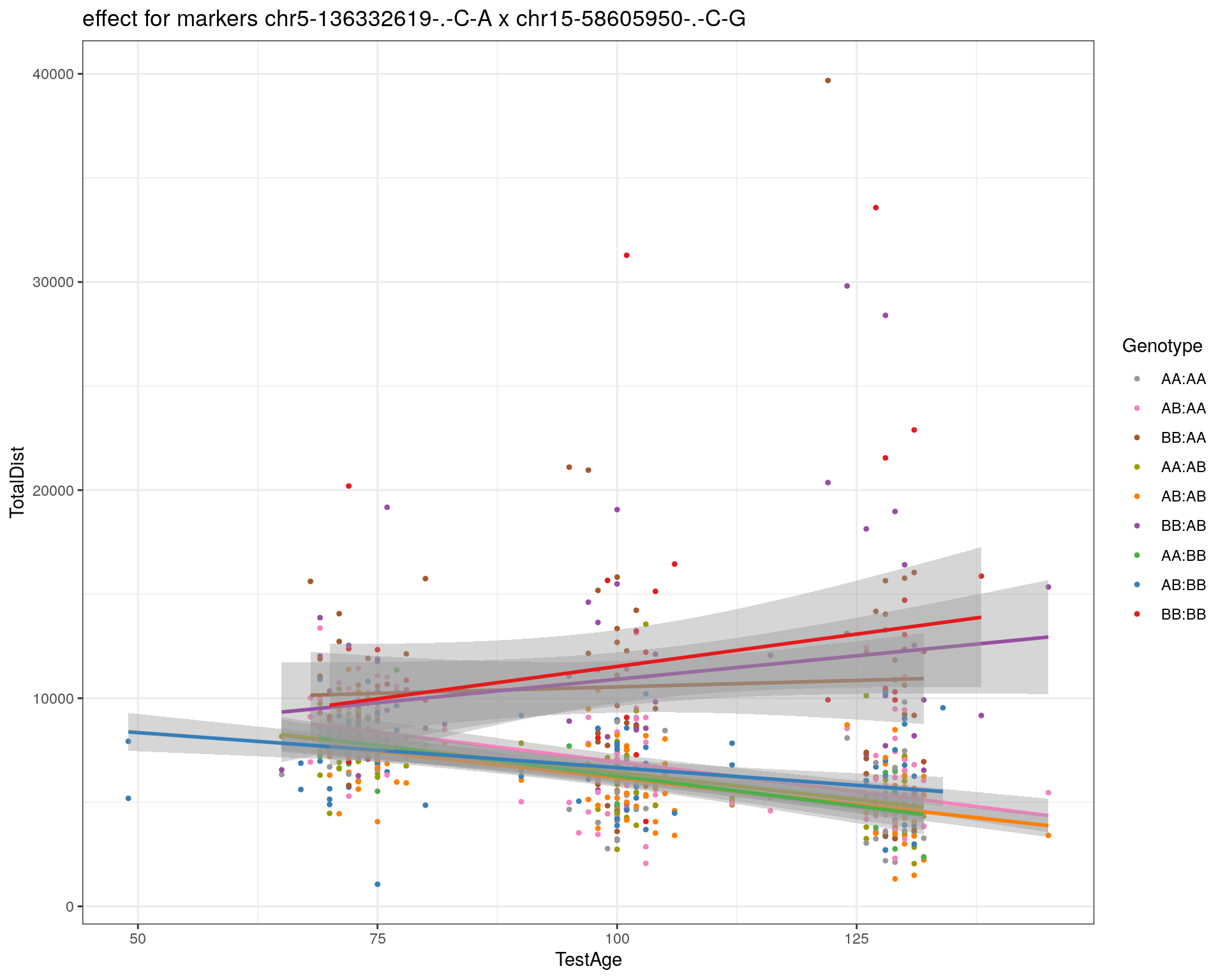
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
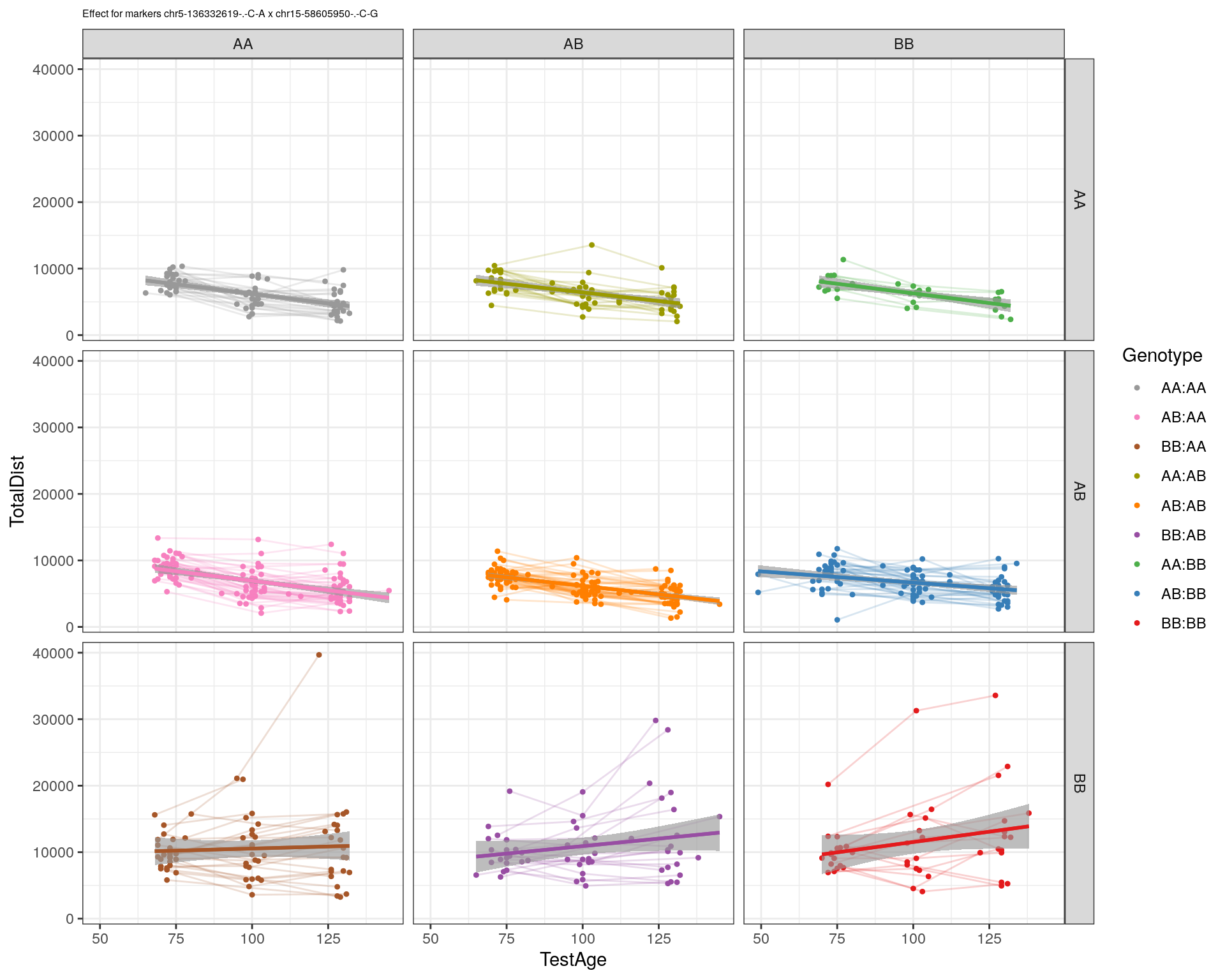
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr15-74727869-.-C-T"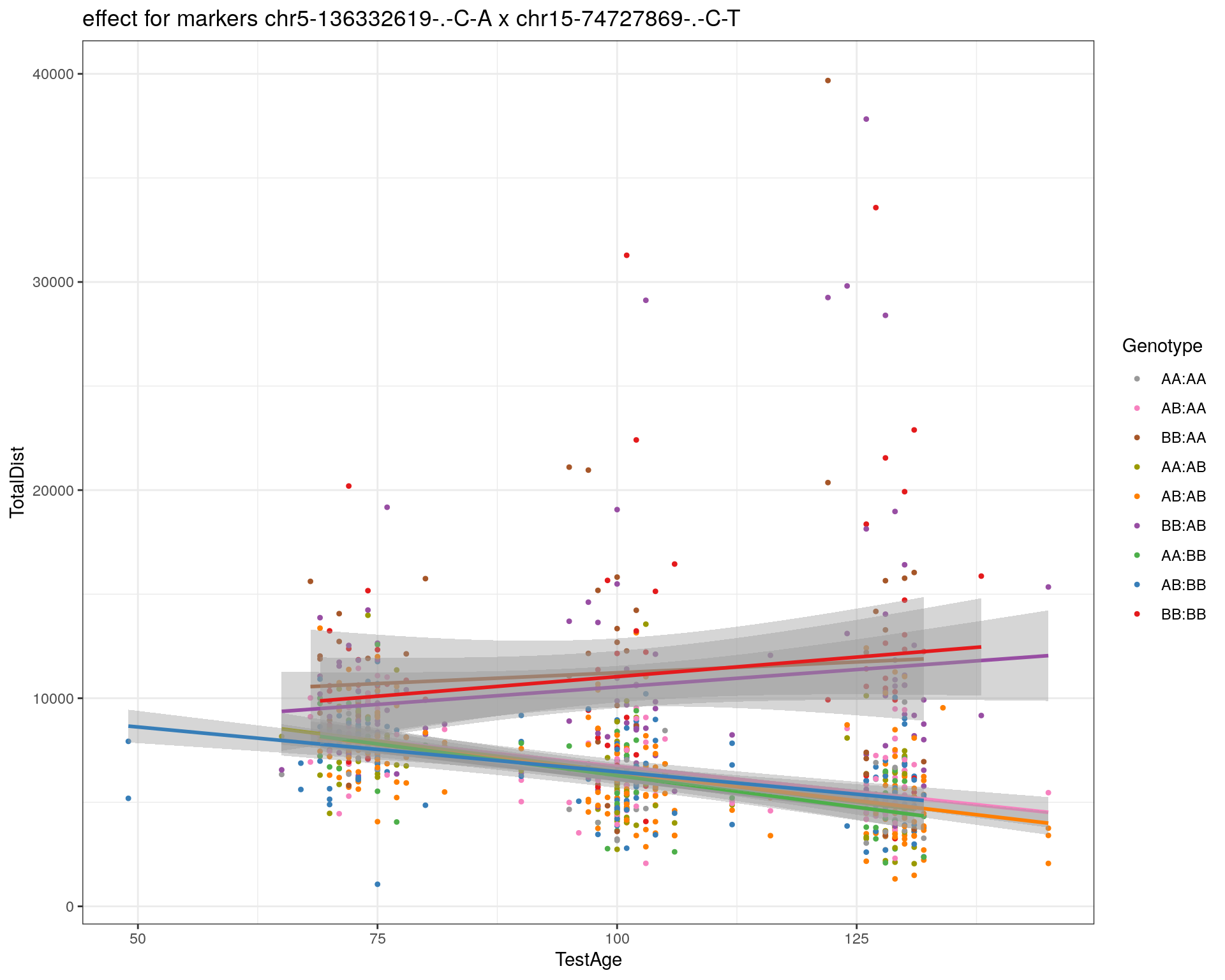
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
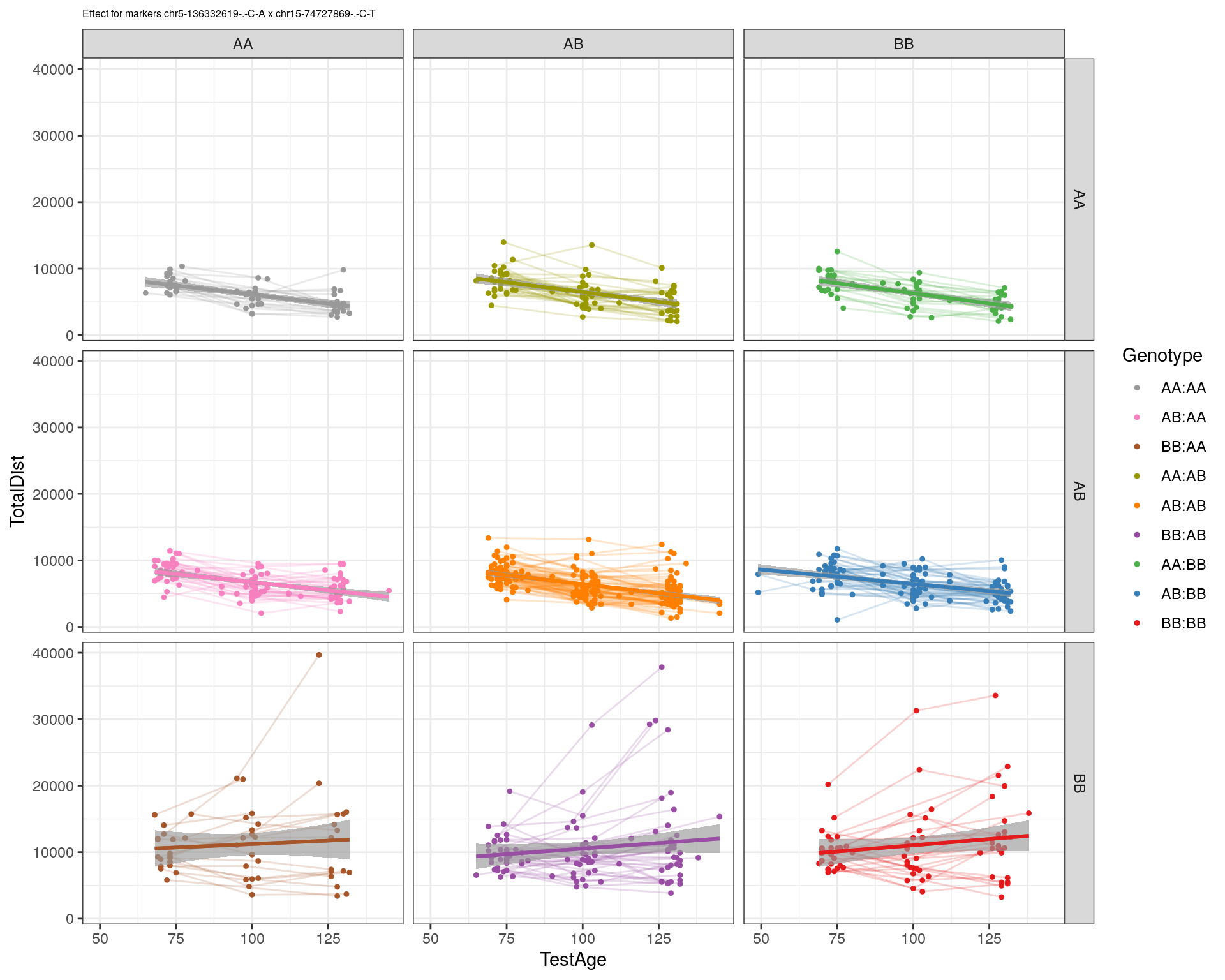
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr16-32593683-.-A-G"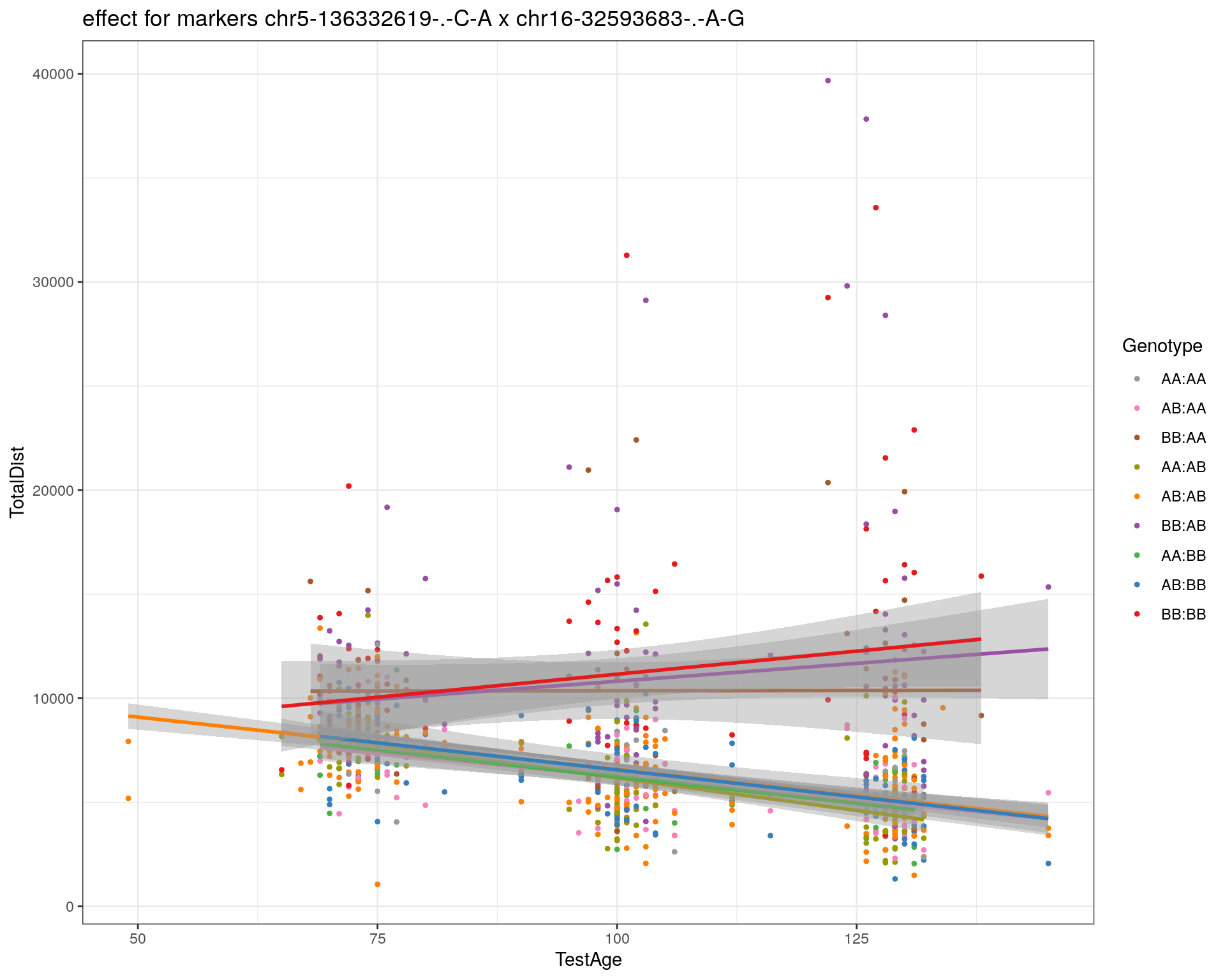
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
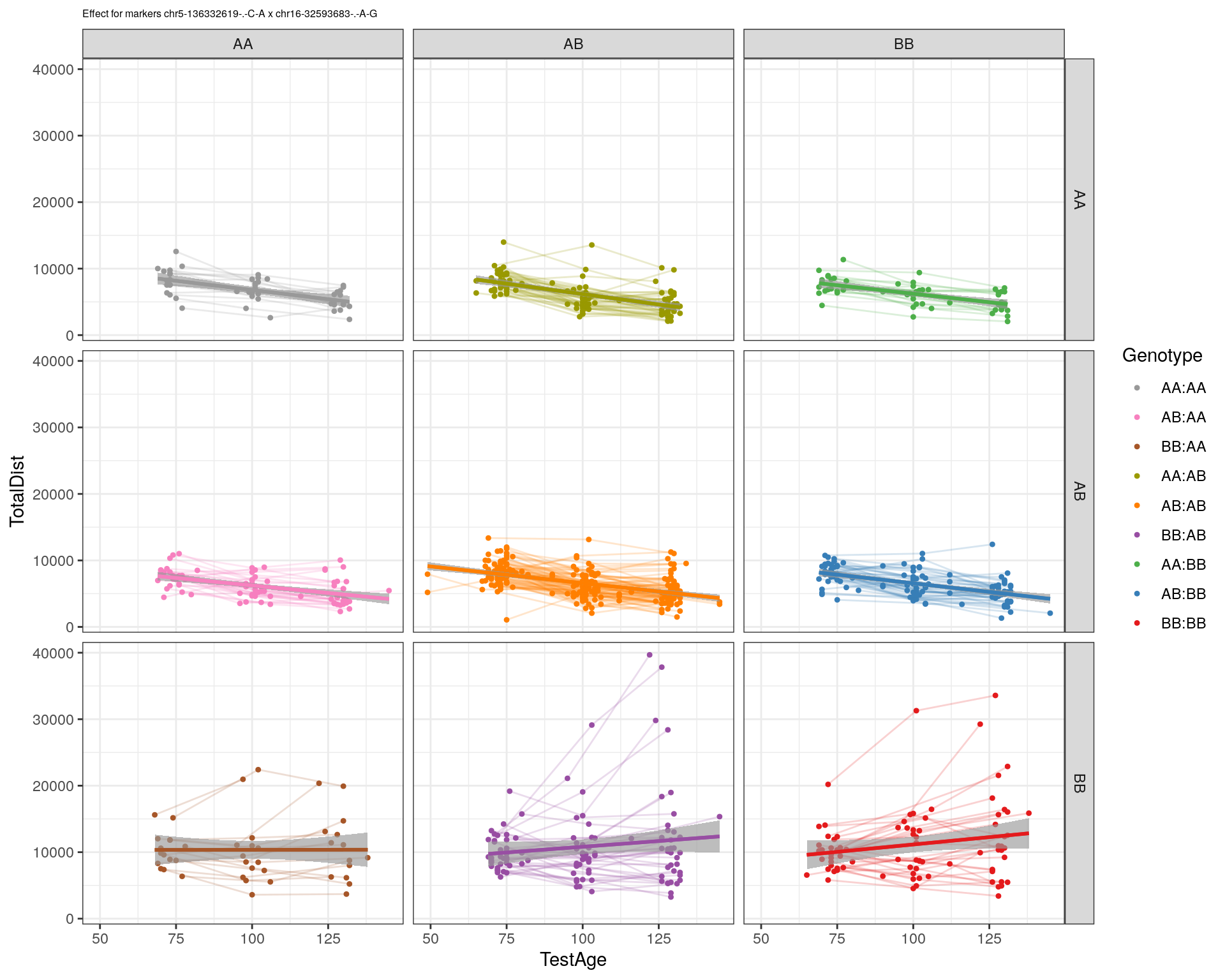
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr16-75648687-.-G-A"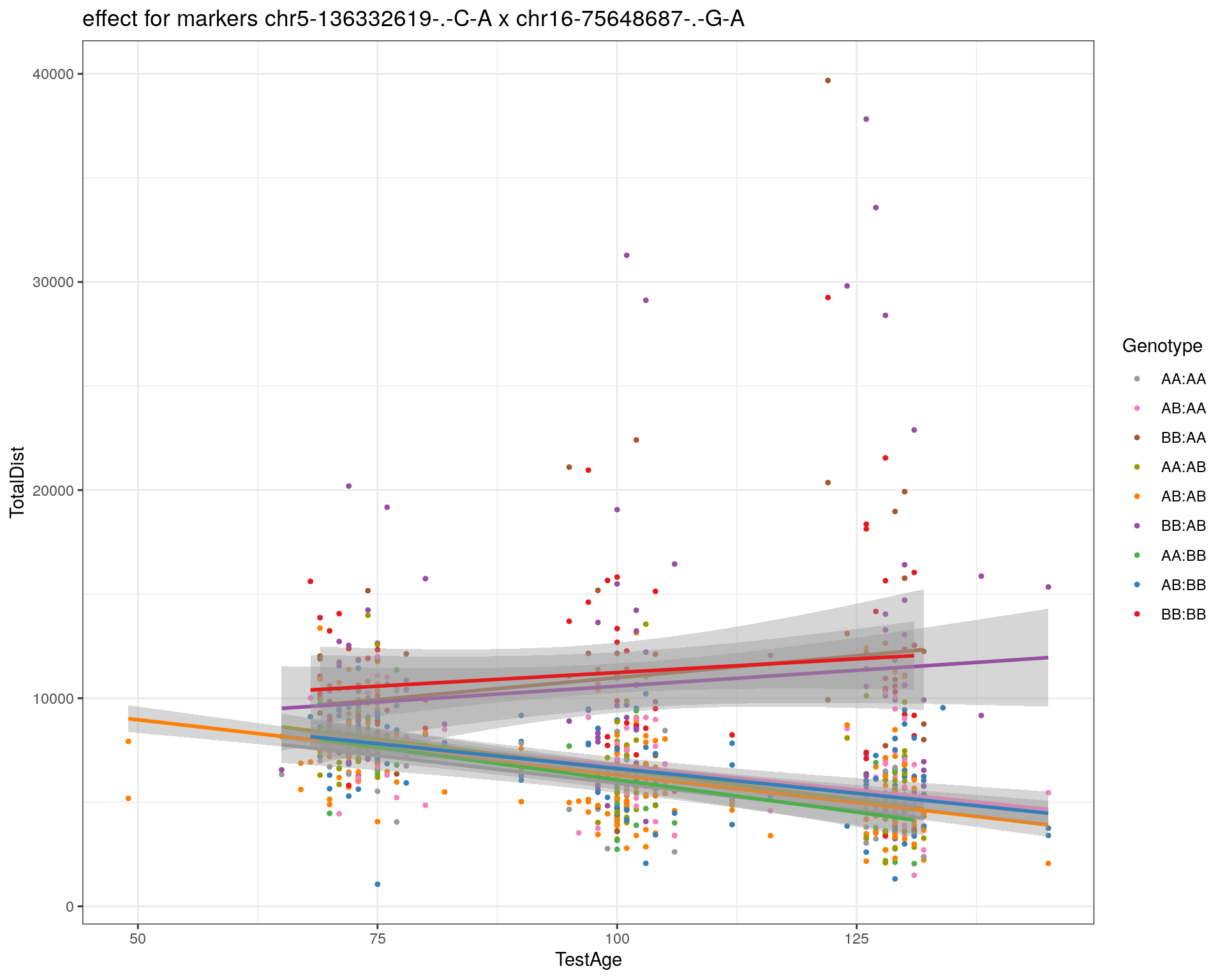
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
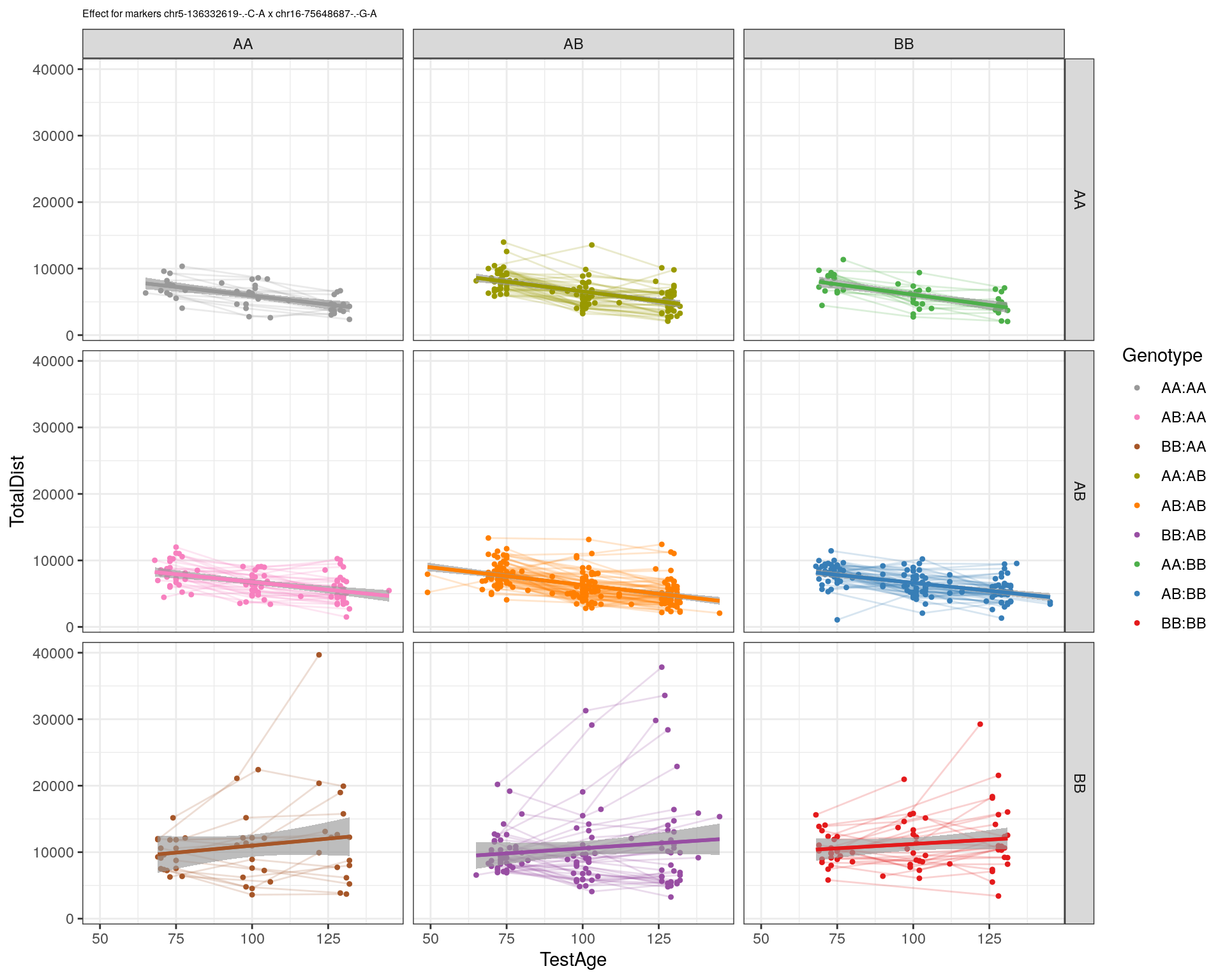
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr17-61305603-.-G-A"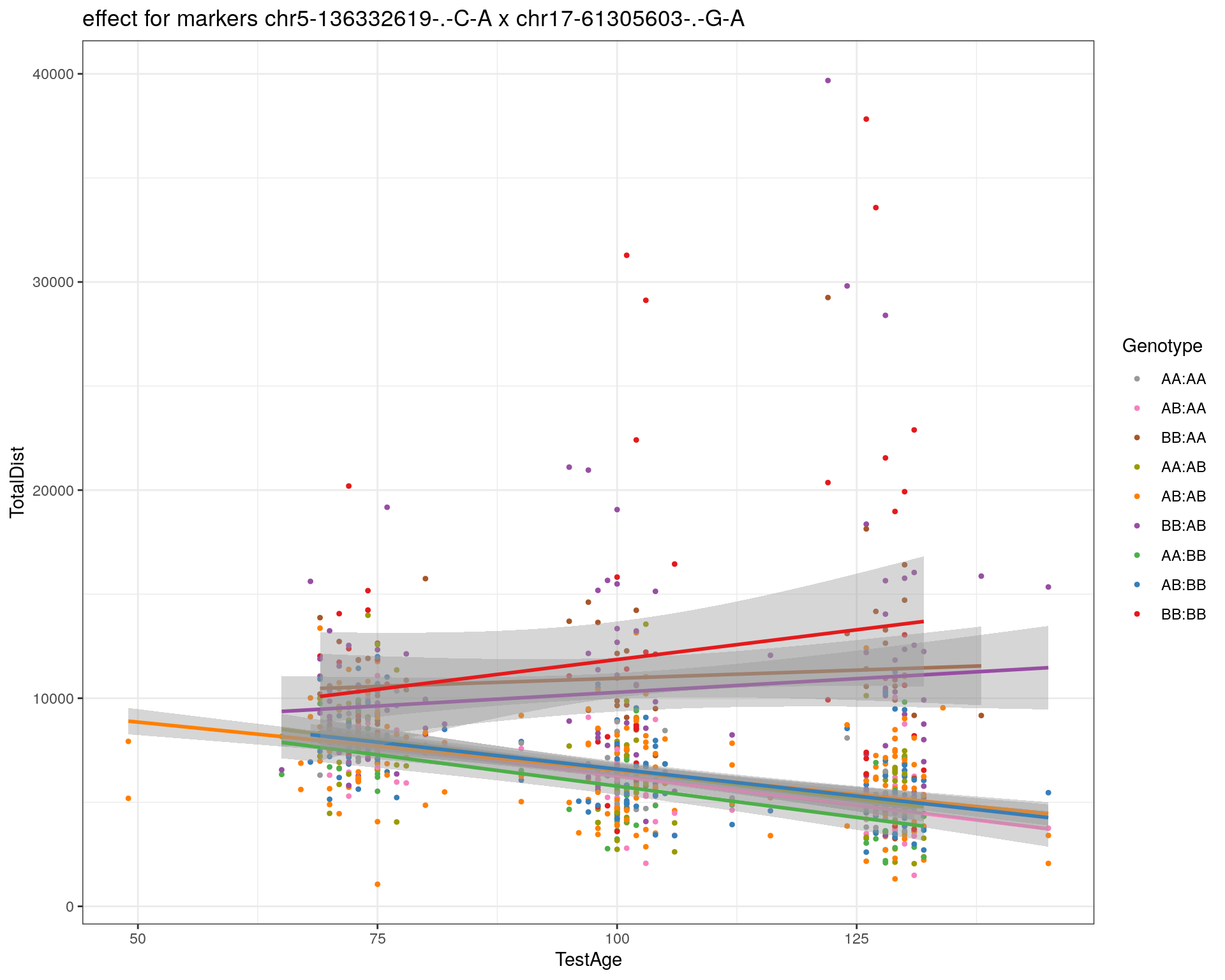
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr17-69219676-.-G-A"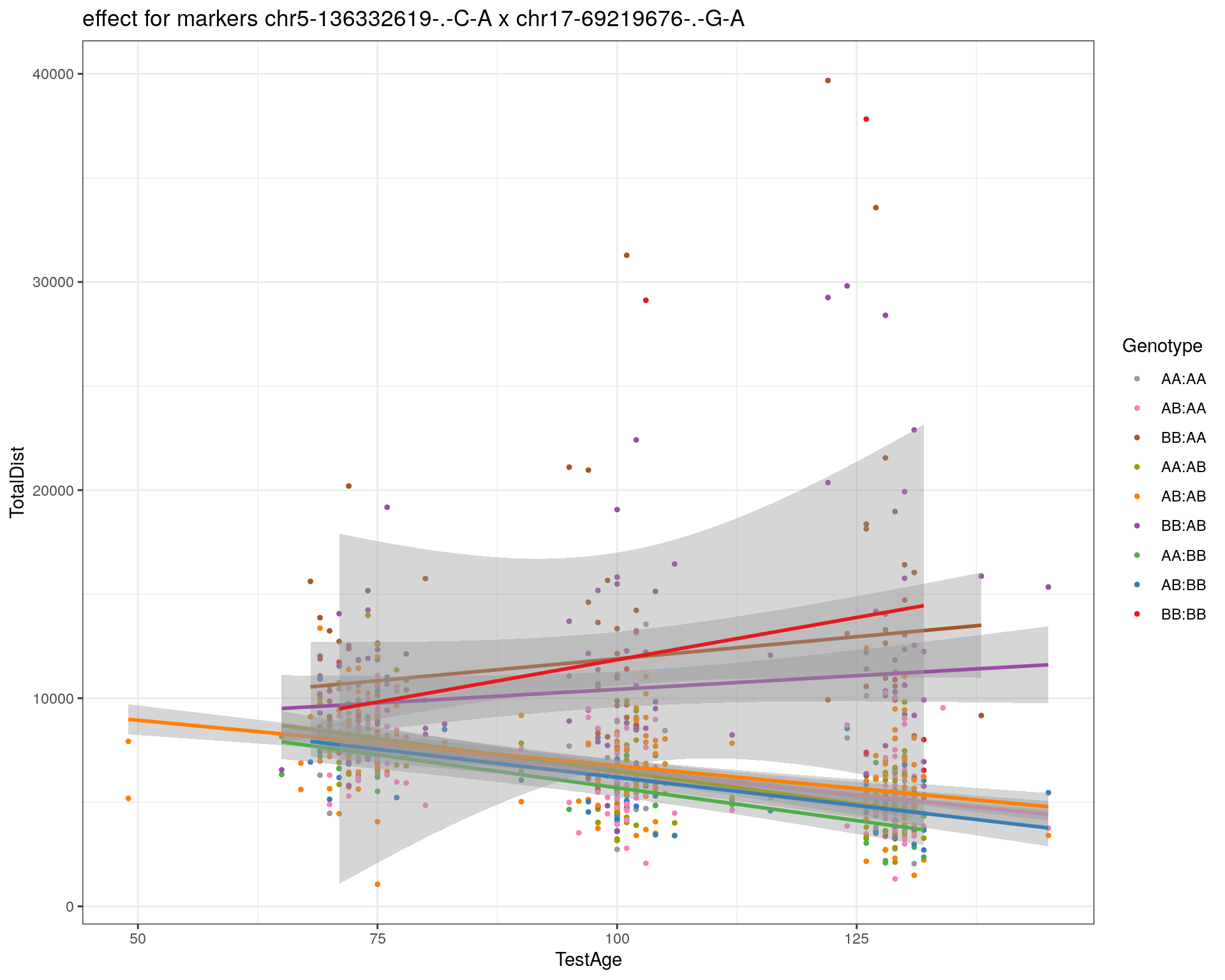
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
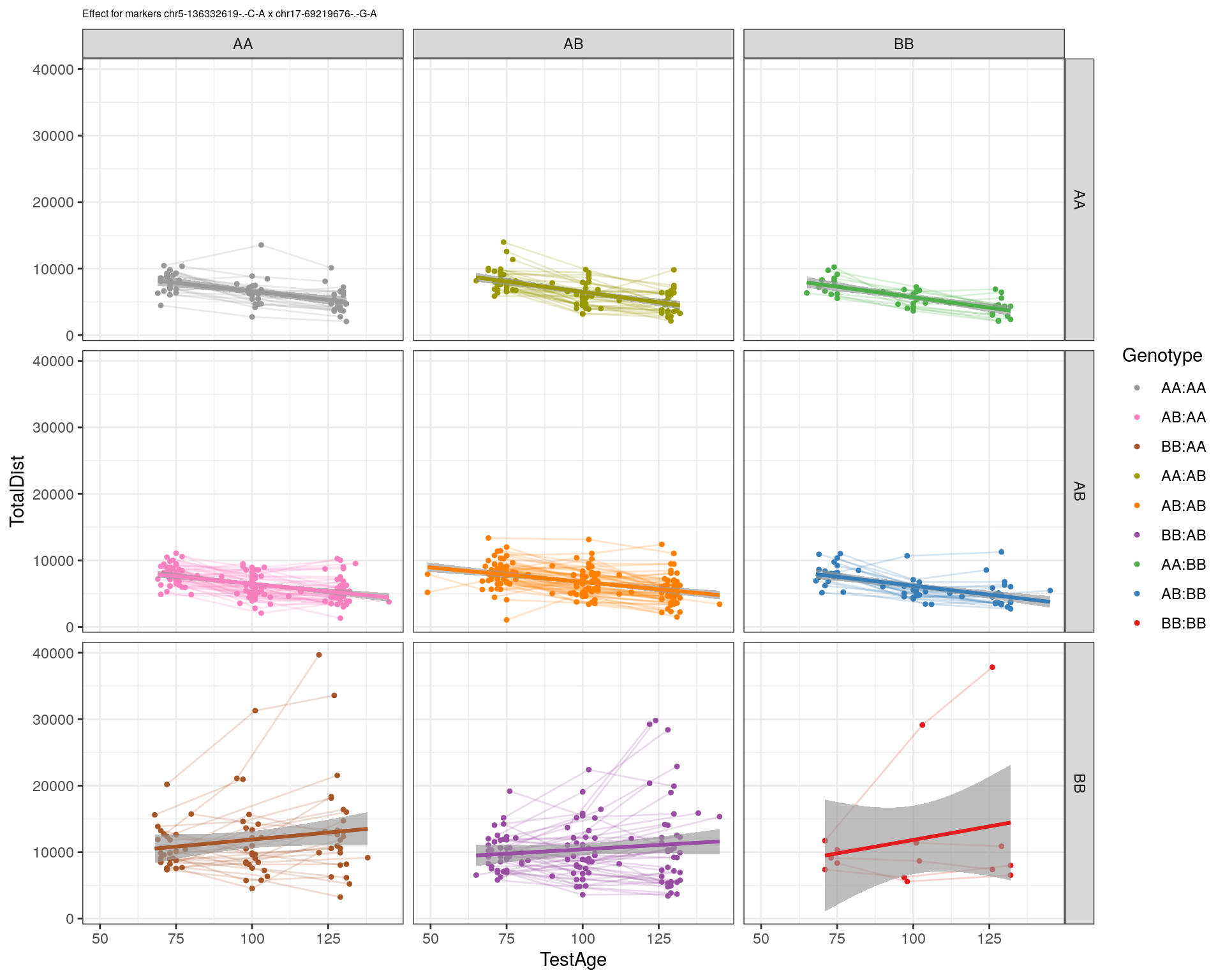
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr18-51300216-.-T-A"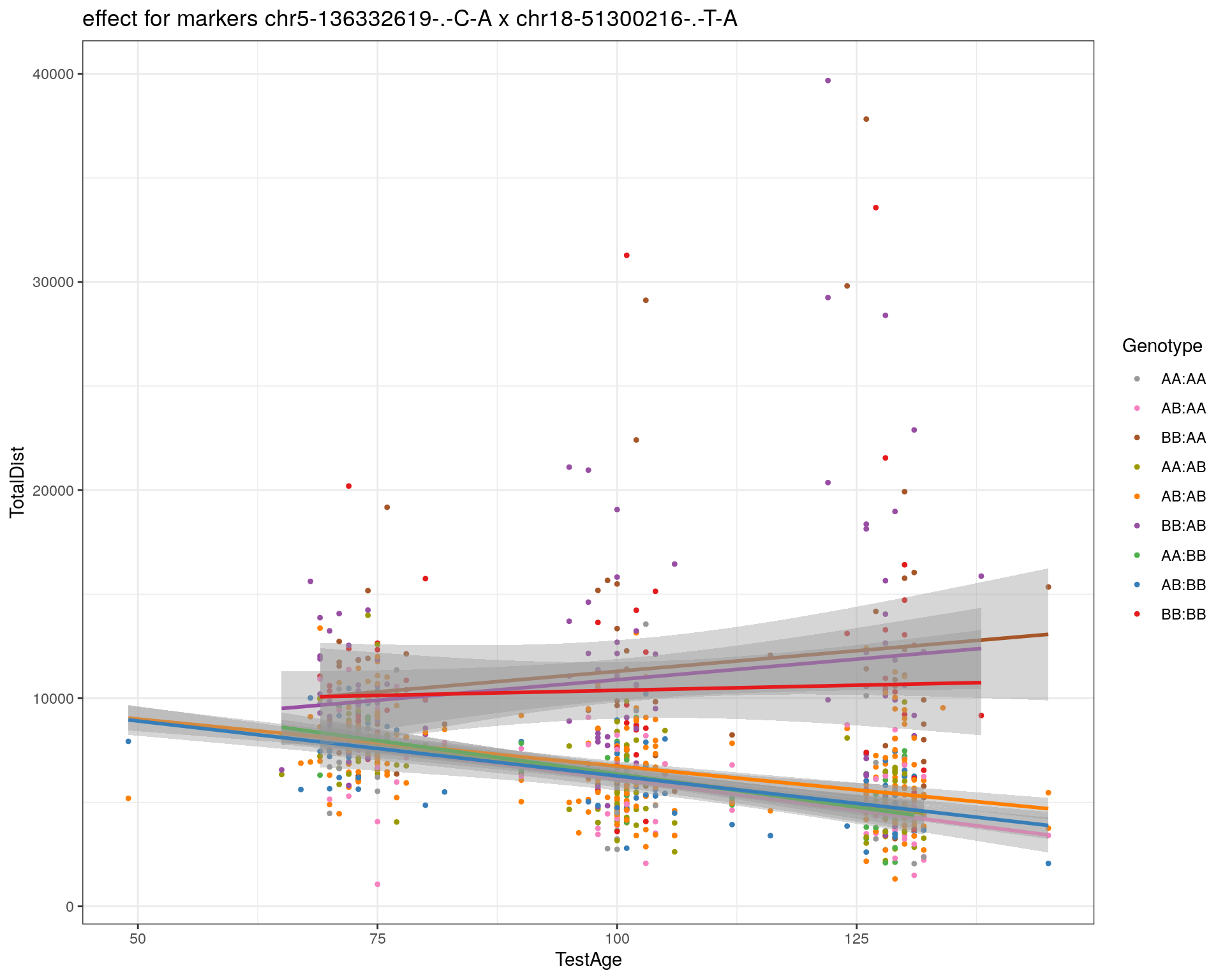
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
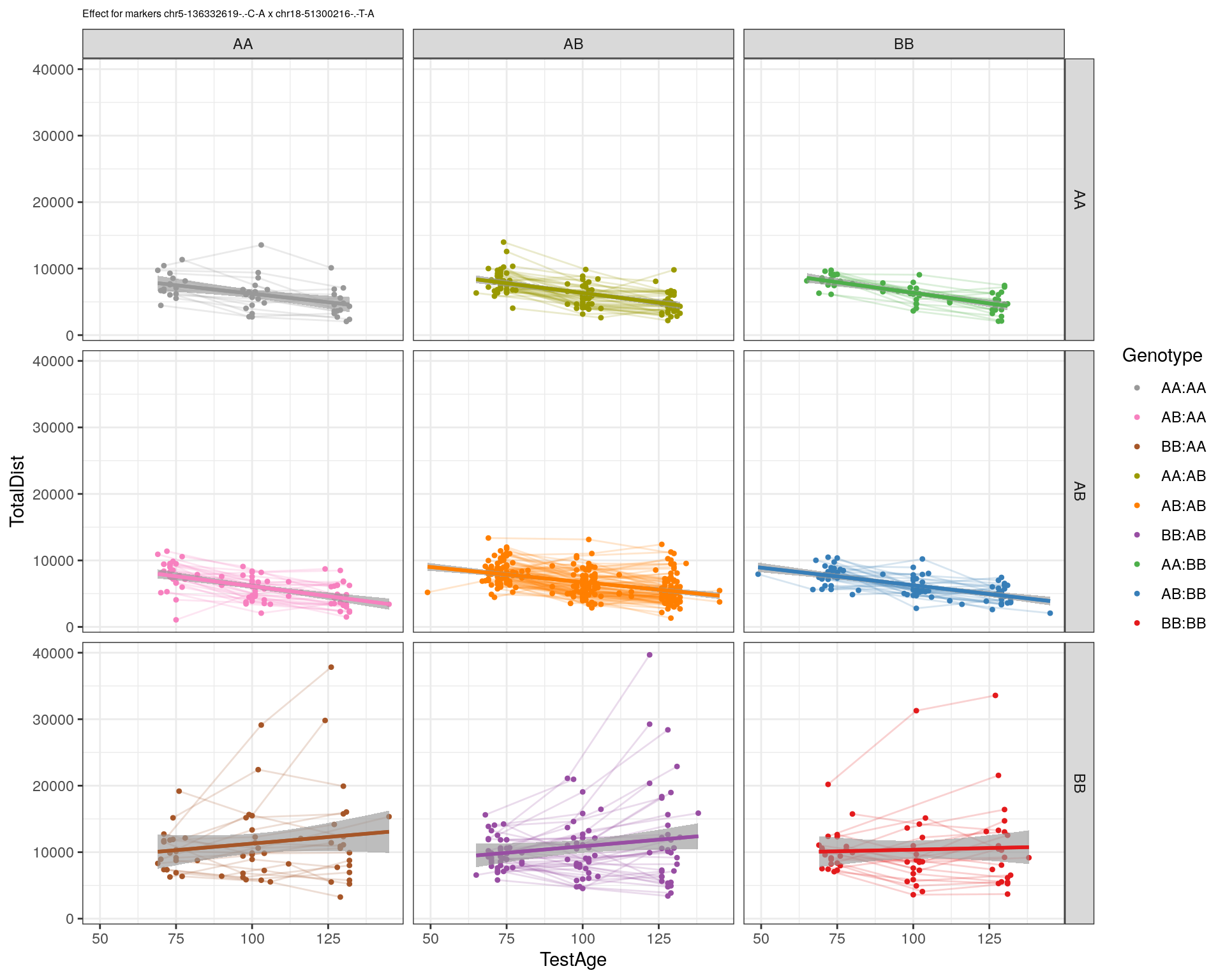
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr18-89347666-.-A-G"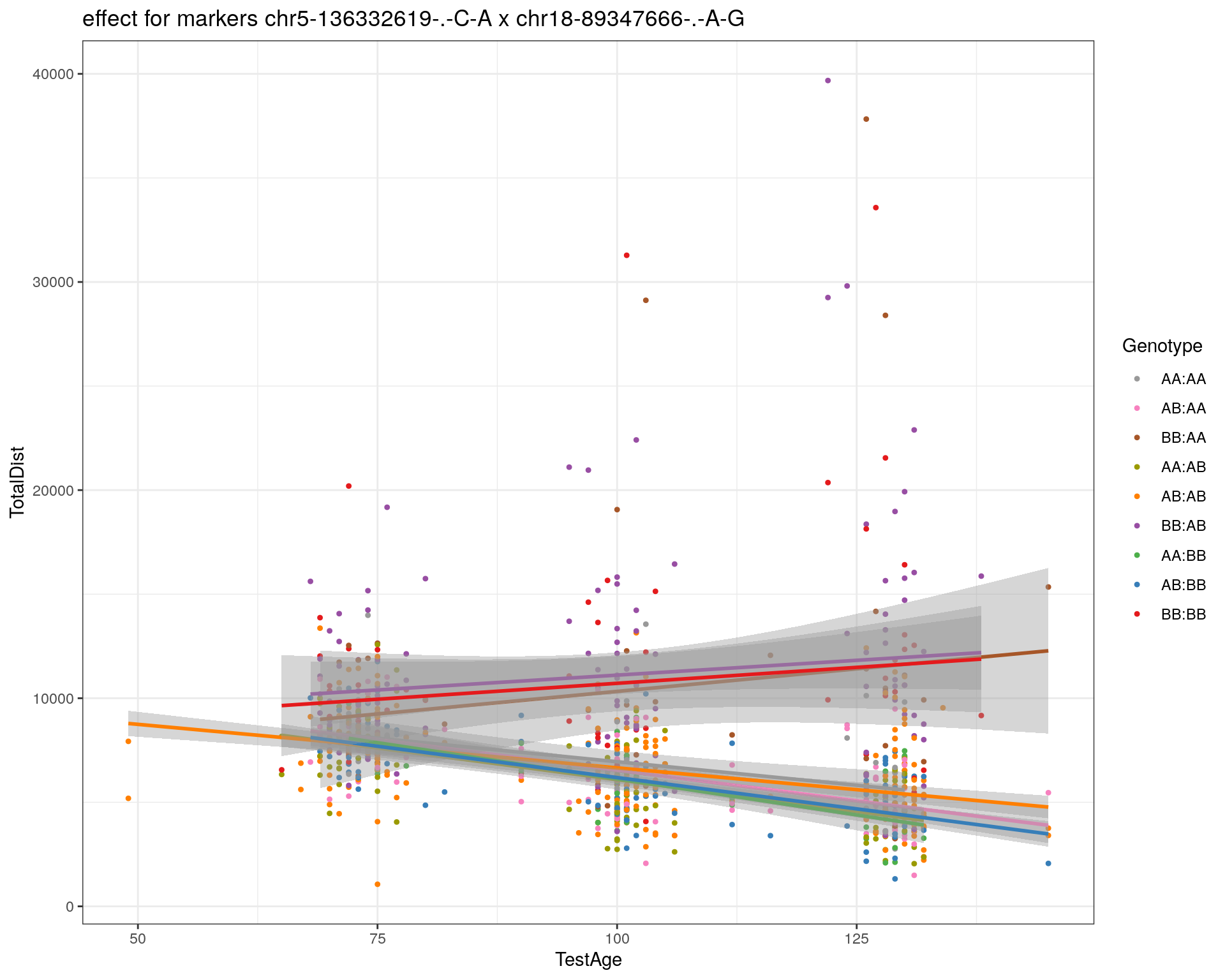
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
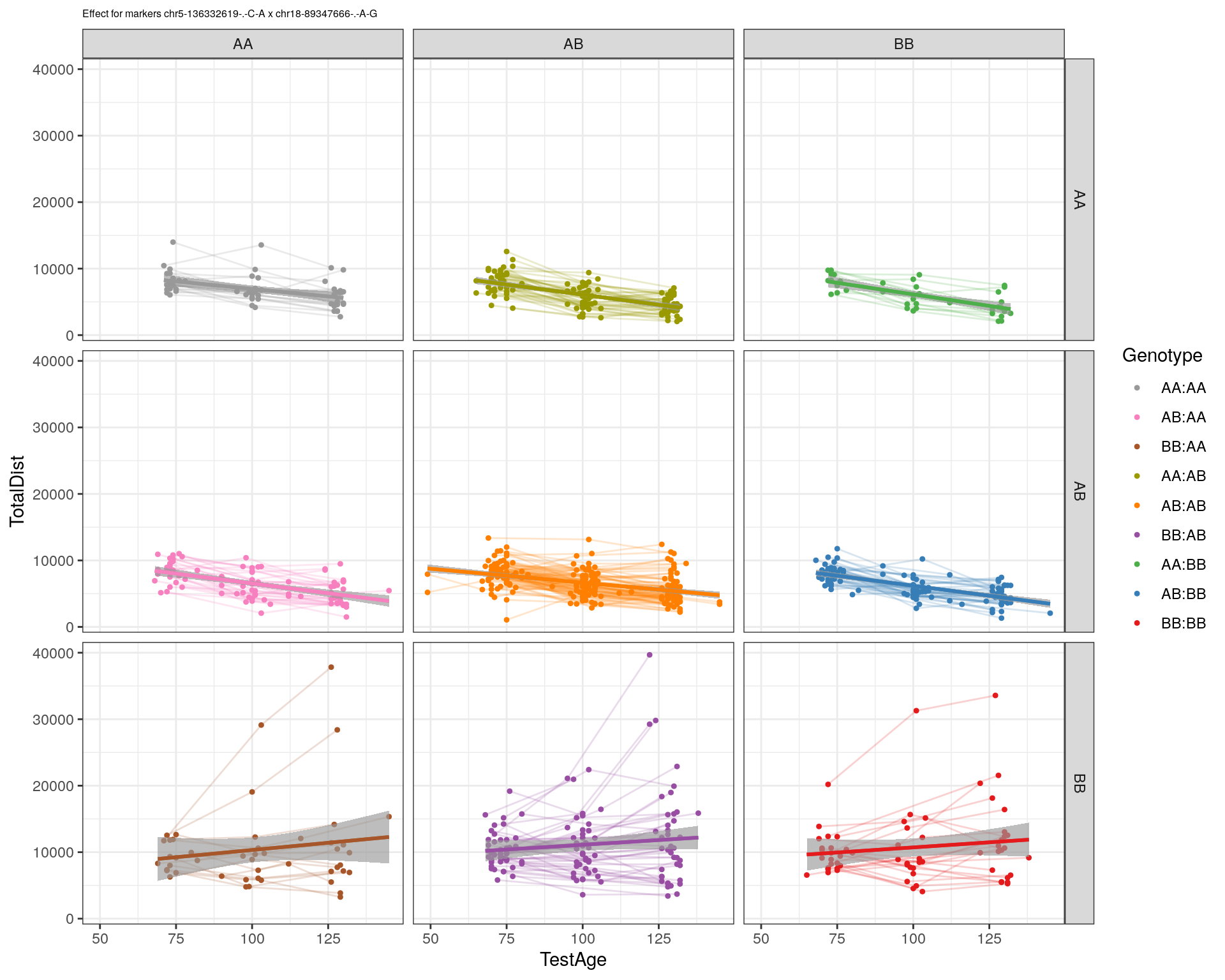
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr19-29815794-.-A-T"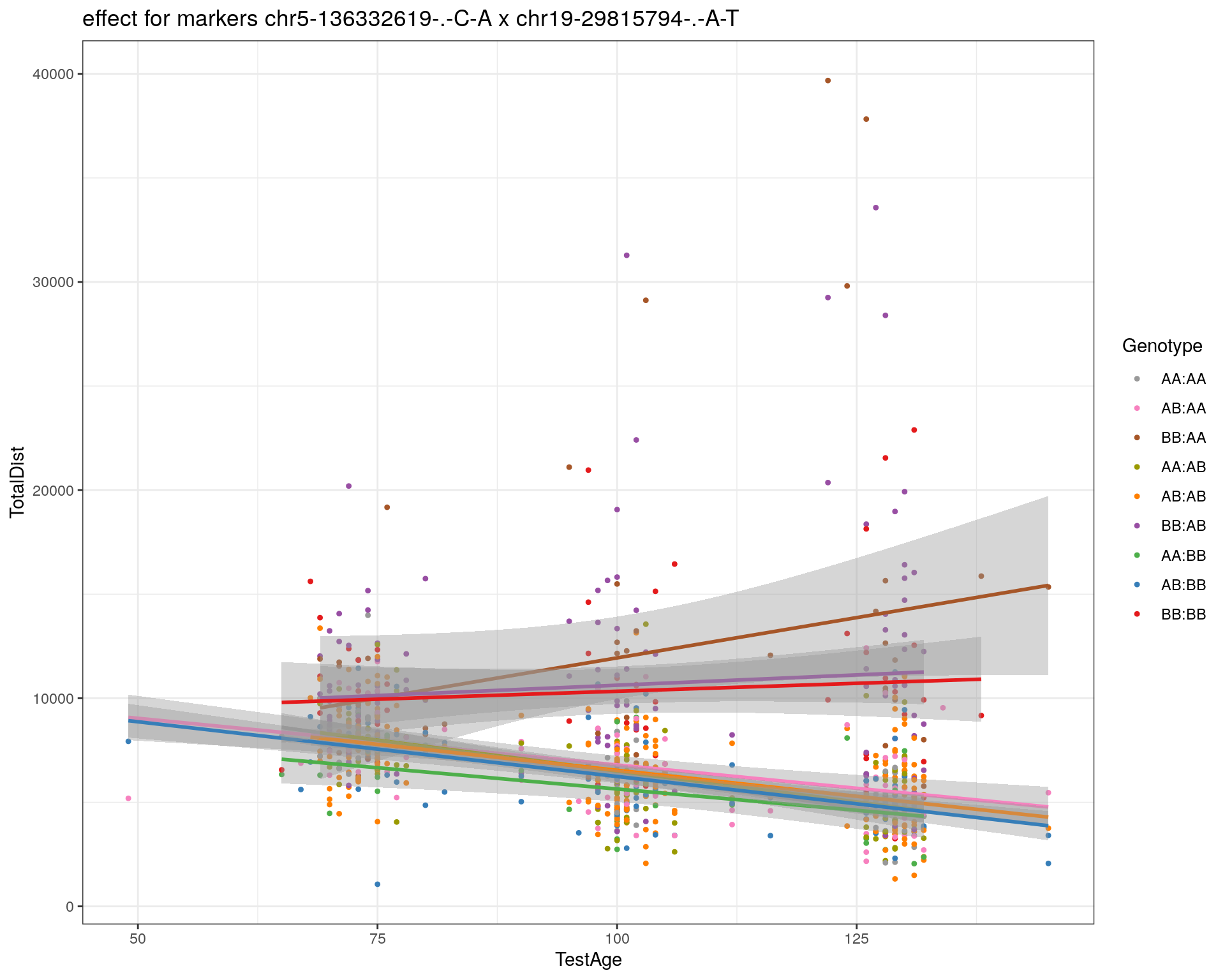
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
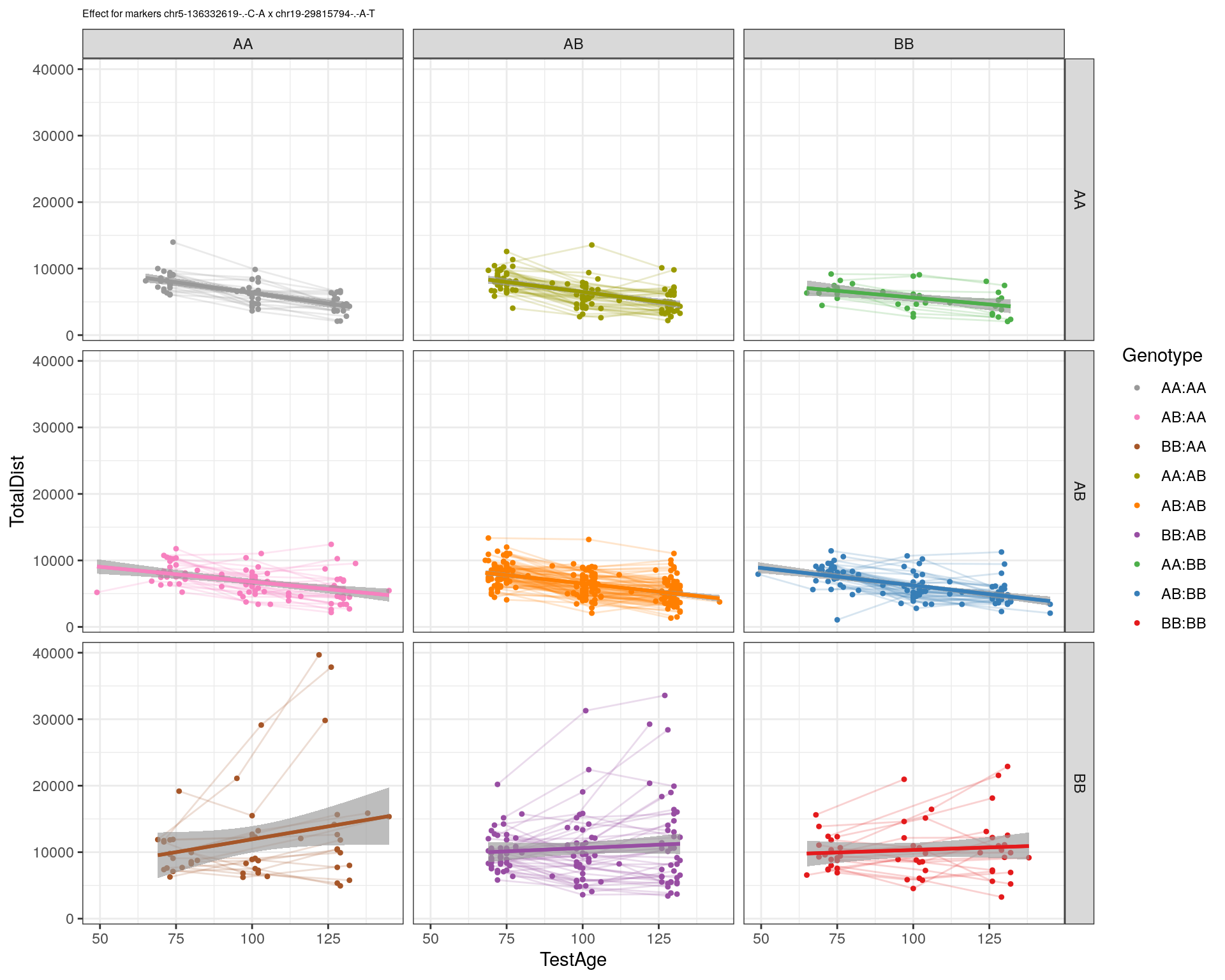
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr19-45958839-.-A-T"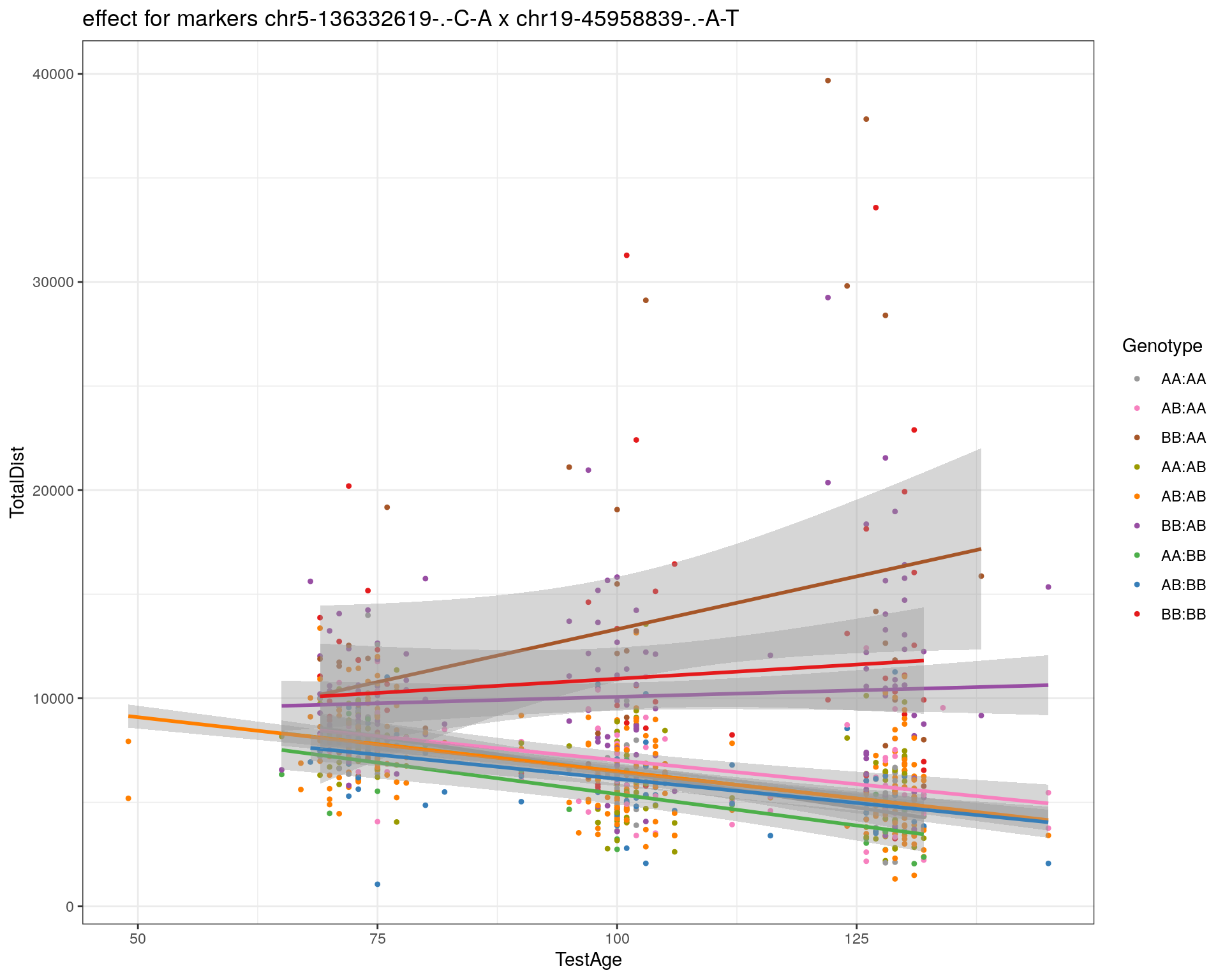
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
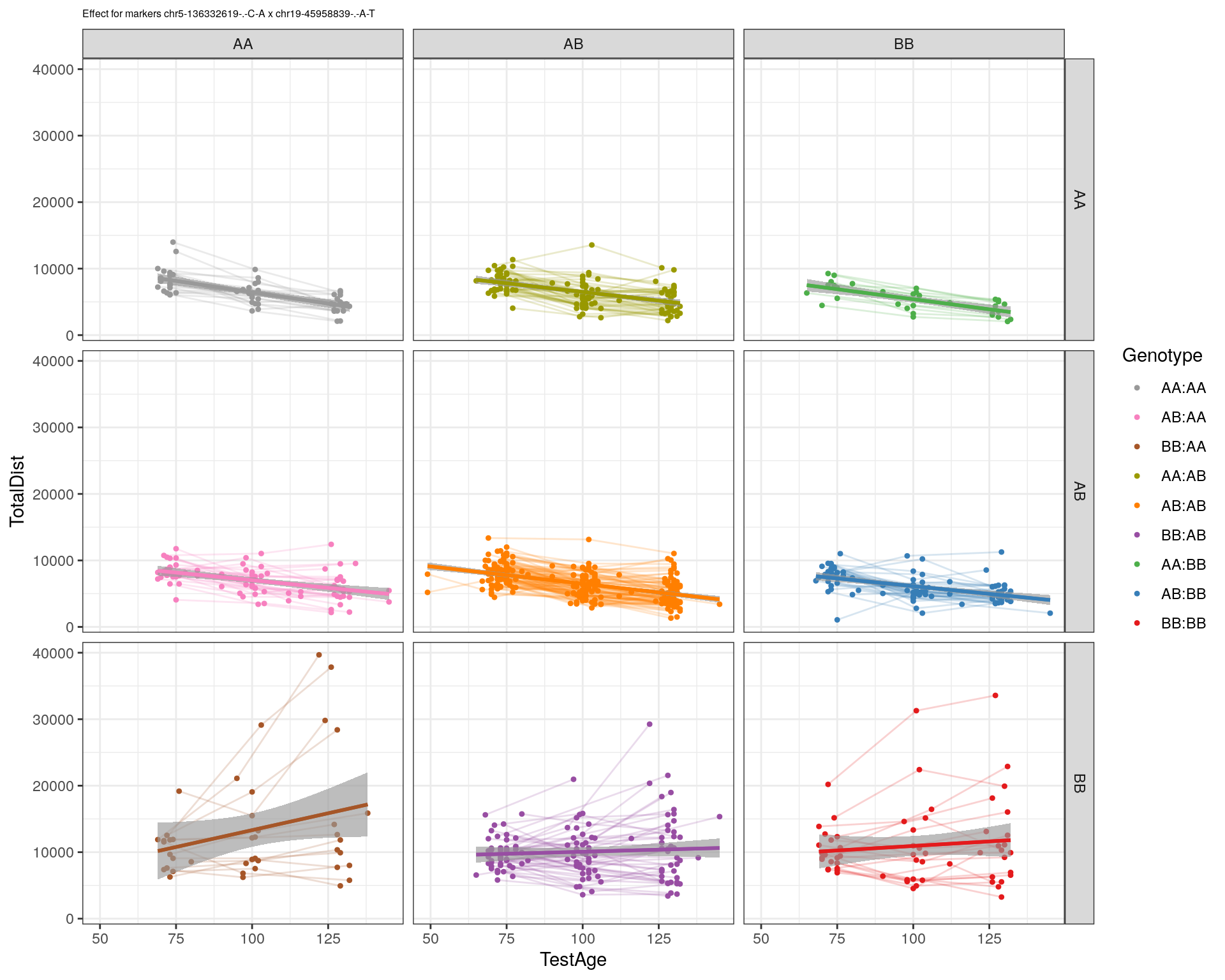
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
[1] "chr19-53469239-.-A-G"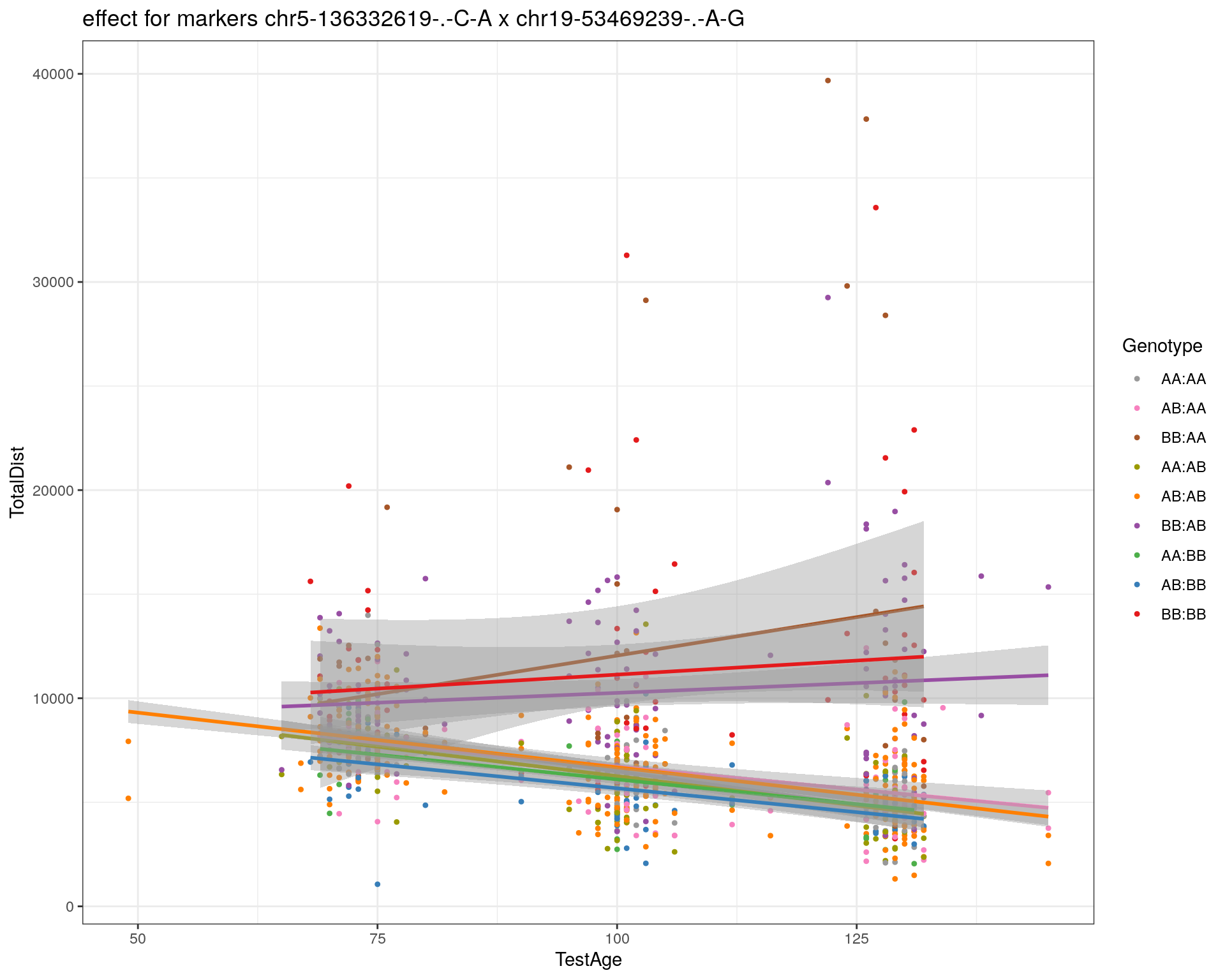
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
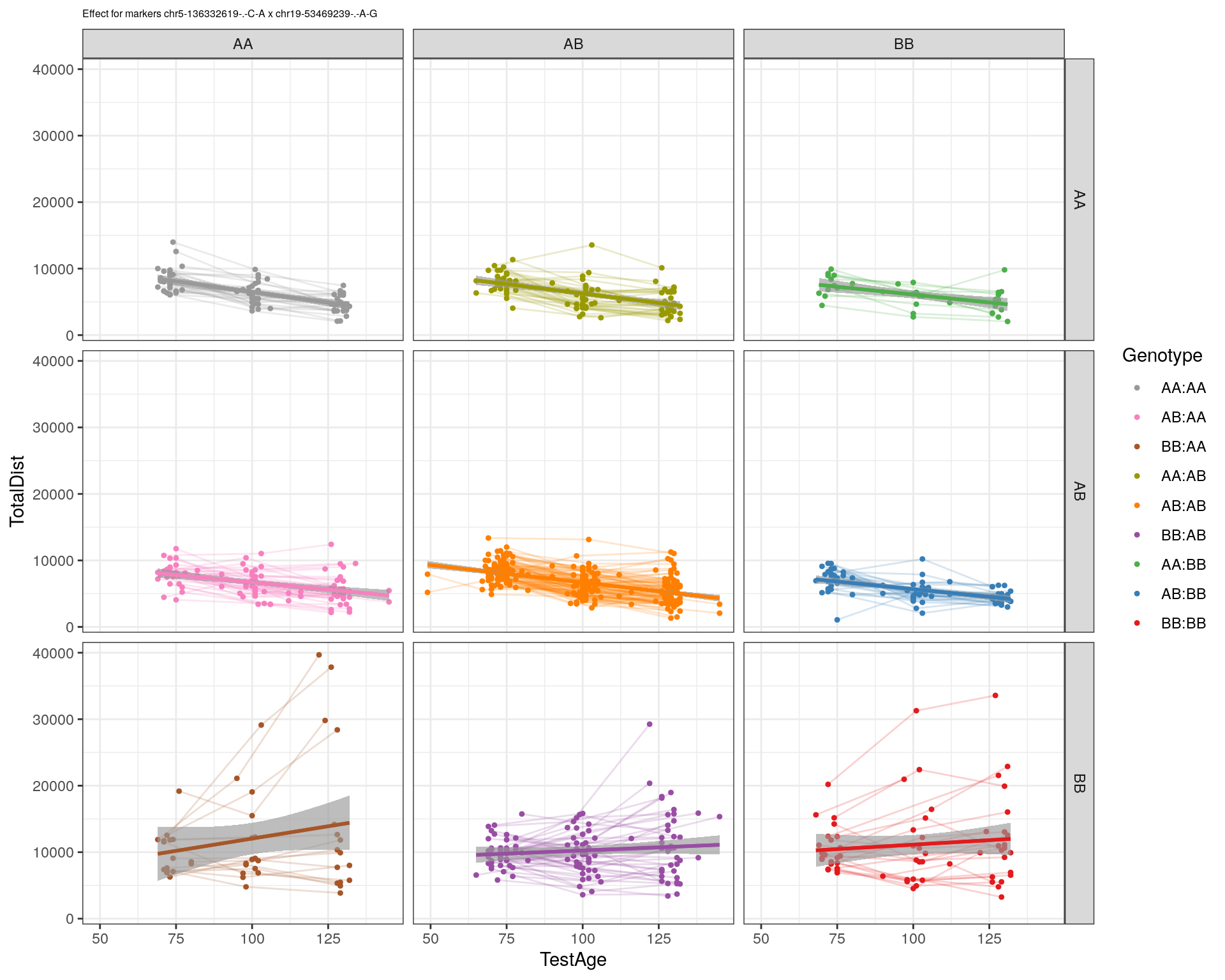
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
pdf(paste0("output/", paste0("WT144_effect_for_marker_", "_int_chr5-136332619-.-C-A"), ".pdf"))
plot_int_graph(WT144, paste0("WT144_effect_for_marker_", "_int_chr5-136332619-.-C-A"), "chr5-136332619-.-C-A")[1] "chr1-65643621-.-C-A"[1] "chr1-74679418-.-A-G"[1] "chr1-79823528-.-A-G"[1] "chr1-86851039-.-A-G"[1] "chr1-188155143-.-G-A"[1] "chr2-7018694-.-G-T"[1] "chr2-79706301-.-T-A"[1] "chr2-92367783-.-T-C"[1] "chr2-95118744-.-T-A"[1] "chr2-102169266-.-C-T"[1] "chr2-130405980-.-A-T"[1] "chr2-137498061-.-T-A"[1] "chr2-145128463-.-A-T"[1] "chr3-59218564-.-T-A"[1] "chr3-98881230-.-G-A"[1] "chr3-106985708-.-A-C"[1] "chr3-135614002-.-T-G"[1] "chr3-145286160-.-C-A"[1] "chr3-146750531-.-A-G"[1] "chr3-152699551-.-G-A"[1] "chr4-32821418-.-C-T"[1] "chr5-75264187-.-T-C"[1] "chr5-90233933-.-A-T"[1] "chr5-97607237-.-T-A"[1] "chr5-107507008-.-C-T"[1] "chr5-112799362-.-C-T"[1] "chr5-120660771-.-T-A"[1] "chr5-122167749-.-T-C"[1] "chr5-125153901-.-C-T"[1] "chr5-126058493-.-T-A"[1] "chr5-126551890-.-T-A"[1] "chr5-127565504-.-T-A"[1] "chr5-130394596-.-T-C"[1] "chr5-130943677-.-T-A"[1] "chr5-131298836-.-T-C"[1] "chr5-133052642-.-T-G"[1] "chr5-135960983-.-T-C"[1] "chr5-136332619-.-C-A"[1] "chr5-136844385-.-T-G"[1] "chr5-138956439-.-T-C"[1] "chr5-139706036-.-T-C"[1] "chr5-139891407-.-T-C"[1] "chr5-140071081-.-C-A"[1] "chr7-35061547-.-A-G"[1] "chr7-44272713-.-T-C"[1] "chr7-66785411-.-A-G"[1] "chr7-112510032-.-A-T"[1] "chr8-91986173-.-T-C"[1] "chr8-99395759-.-T-C"[1] "chr8-112465004-.-G-A"[1] "chr9-64698947-.-C-T"[1] "chr9-65857475-.-T-C"[1] "chr9-69415567-.-T-A"[1] "chr9-71739608-.-T-A"[1] "chr9-80465702-.-T-A"[1] "chr10-24134777-.-A-T"[1] "chr11-96730656-.-A-G"[1] "chr12-43136399-.-A-T"[1] "chr12-69197484-.-C-A"[1] "chr12-86851130-.-T-A"[1] "chr12-112415100-.-G-A"[1] "chr13-20165684-.-T-A"[1] "chr13-115031098-.-T-C"[1] "chr14-11709591-.-T-C"[1] "chr14-50642778-.-G-A"[1] "chr14-62407765-.-T-A"[1] "chr14-69691477-.-G-T"[1] "chr15-58605950-.-C-G"[1] "chr15-74727869-.-C-T"[1] "chr16-32593683-.-A-G"[1] "chr16-75648687-.-G-A"[1] "chr17-61305603-.-G-A"[1] "chr17-69219676-.-G-A"[1] "chr18-51300216-.-T-A"[1] "chr18-89347666-.-A-G"[1] "chr19-29815794-.-A-T"[1] "chr19-45958839-.-A-T"[1] "chr19-53469239-.-A-G"dev.off()png
2 Figure panel for publication
# define a function that emits the desired plot
figA <- function() {
par(
mar = c(3, 5, 1, 1),
mgp = c(2, 1, 0)
)
#plot phenotypes: slope and totaldistance3
plot(out.mr[[2]], col= "blue", ylim = c(0, 30), ylab = "LOD score")
plot(out.mr[[1]], col= "green", add=TRUE)
abline(h=3.2, col="red", lty=2, lwd=3)
# Add a legend
legend("topright",
legend=c("Slope", "Total distance traveled"),
col=c("blue", "green"), lty=1, cex=0.8)
}
figB <- function() {
par(
mar = c(3, 5, 1, 0.5),
mgp = c(2, 1, 0)
)
#only at chr 5 and 14
#plot phenotypes: slope and totaldistance3
plot(out.mr[[2]], col= "blue", ylim = c(0, 30), ylab = "LOD score", chr = c(5), main = "QTL Interval Chr 5")
plot(out.mr[[1]], col= "green", add=TRUE, chr = c(5))
abline(h=3.2, col="red", lty=2, lwd=3)
#abline(v= 76.0, col="black", lty=2, lwd=1)
text(75.5, 29, "5@76.0", cex = 0.75)
segments(x0 = 76,
x1 = 76,
y0 = 0,
y1 = 28,
col="black", lty=2, lwd=1)
segments(x0 = 72.40574,
x1 = 77.76853,
y0 = 2,
y1 = 2,
col="black", lty=1, lwd=2)
segments(x0 = 72.40574,
x1 = 72.40574,
y0 = 1.85,
y1 = 2.15,
col="black", lty=1, lwd=2)
segments(x0 = 77.76853,
x1 = 77.76853,
y0 = 1.85,
y1 = 2.15,
col="black", lty=1, lwd=2)
text(63, 1, "1.5-LOD interval", cex = 0.75)
# Add a legend
# legend("topright",
# legend=c("Slope", "Total Distance traveled"),
# col=c("blue", "green"), lty=1, cex=0.8)
}
figC <- function() {
par(
mar = c(3, 5, 1, 0.5),
mgp = c(2, 1, 0)
)
#only at chr 5 and 14
#plot phenotypes: slope and totaldistance3
plot(out.mr[[2]], col= "blue", ylim = c(0, 10), ylab = "LOD score", chr = c(14), main = "QTL Interval Chr 14")
plot(out.mr[[1]], col= "green", add=TRUE, chr = c(14))
abline(h=3.2, col="red", lty=2, lwd=3)
#abline(v= 76.0, col="black", lty=2, lwd=1)
text(34, 7, "14@36.1", cex = 0.75)
segments(x0 = 36.07219,
x1 = 36.07219,
y0 = 0,
y1 = 6.4,
col="black", lty=2, lwd=1)
segments(x0 = 26.14,
x1 = 36.07219,
y0 = 2,
y1 = 2,
col="black", lty=1, lwd=2)
segments(x0 = 26.14,
x1 = 26.14,
y0 = 1.85,
y1 = 2.15,
col="black", lty=1, lwd=2)
segments(x0 = 36.07219,
x1 = 36.07219,
y0 = 1.85,
y1 = 2.15,
col="black", lty=1, lwd=2)
text(29.5, 1, "1.5-LOD interval", cex = 0.75)
# Add a legend
# legend("topright",
# legend=c("Slope", "Total Distance traveled"),
# col=c("blue", "green"), lty=1, cex=0.8)
}
figD <- function() {
par(mar = c(5, 5, 1, 0.5),
mgp = c(3, 0.8, 0))
#interaction_effect for slope
effectplot(WT144, main = "",
mname1 = marker[[1]],
mname2 = marker[[2]], ylab = "Total distance traveled",
xlab = marker[[2]],
pheno.col = 11, add.legend=F)
# Add a legend
legend("topleft", legend=c("AA", "AB", "BB"), title = marker[[1]],
col=c("black","red", "blue"), pch = 1, lty=1, cex=0.8)
}
figE <- function() {
par(
mar = c(3, 5, 1, 1),
mgp = c(3, 1, 0)
)
#interaction_effect for slope
effectplot(WT144, main = "",
mname1 = marker[[1]],
mname2 = marker[[2]], ylab = "Slope",xlab = marker[[2]],
pheno.col = 15, add.legend=F)
}
cross = WT144
basem = "chr5-136332619-.-C-A"
# Plot the Dist vs age, separate by genotype for each marker
allphen <- cross$pheno
allgeno <- as.data.frame(pull.geno(cross))
rownames(allgeno) <- cross$pheno$AnimalName
allphen <- merge(allphen, allgeno, by.x="AnimalName", by.y="row.names", all.x=TRUE)
#pdf(paste0("output/", name, ".pdf"))
#for (mar in colnames(allgeno)){
mar = "chr14-69691477-.-G-T"
print(mar)[1] "chr14-69691477-.-G-T" subplot <- allphen[!is.na(allphen[,mar, drop=FALSE]),] %>%
pivot_longer(cols = starts_with("TestAge_"), names_to="test.no", values_to = "Age") %>%
separate(test.no, c("empty1", "test1")) %>%
pivot_longer(cols = starts_with("TotalDist_"), names_to="test.no.2", values_to = "Dist") %>%
separate(test.no.2, c("empty2", "test2")) %>% filter(test1==test2)
subplot <- as.data.frame(subplot)
subplot$interactions <- factor(subplot[, basem] + subplot[, mar]*3)
# 1:1 -> 4 1:2 -> 7 1:3 -> 10
# 2:1 -> 5 2:2 -> 8 2:3 -> 11
# 3:1 -> 6 3:2 -> 9 3:3 -> 12
itypes <- c(1,2,3,"AA:AA", "AB:AA", "BB:AA",
"AA:AB", "AB:AB", "BB:AB",
"AA:BB", "AB:BB", "BB:BB")
levels(subplot$interactions) <- itypes[as.integer(levels(subplot$interactions))]
#replaces 1 2 and 3 as AA, AB and BB
subplot[, basem][subplot[, basem] == 1] <- "AA"
subplot[, basem][subplot[, basem] == 2] <- "AB"
subplot[, basem][subplot[, basem] == 3] <- "BB"
subplot[, mar][subplot[, mar] == 1] <- "AA"
subplot[, mar][subplot[, mar] == 2] <- "AB"
subplot[, mar][subplot[, mar] == 3] <- "BB"
p1 <- ggplot(subplot, aes(Age, Dist, color=interactions, group=interactions)) +
geom_point(shape = 20, size=0.75) +
#scale_color_discrete("Genotype") +
labs(color = "Genotype") +
ylab("TotalDist") +
xlab("Test Age") +
#scale_color_brewer(palette = "Set1") +
scale_color_manual(breaks = c("AA:AA", "AB:AA", "BB:AA", "AA:AB",
"AB:AB", "BB:AB", "AA:BB", "AB:BB", "BB:BB"),
#values = RColorBrewer::brewer.pal(9, "Set1")[9:1]) +
values = c("#999999", "#F781BF", "#A65628","#9a9a00", "#FF7F00", "#984EA3", "#4DAF4A", "#377EB8", "#E41A1C")) +
stat_smooth(method="lm", show.legend = FALSE, formula = y~x) +
labs(title=paste0("effect for markers ", basem, " x ", mar)) +
theme_bw()
print(p1)Warning: Removed 29 rows containing non-finite values (stat_smooth).Warning: Removed 29 rows containing missing values (geom_point).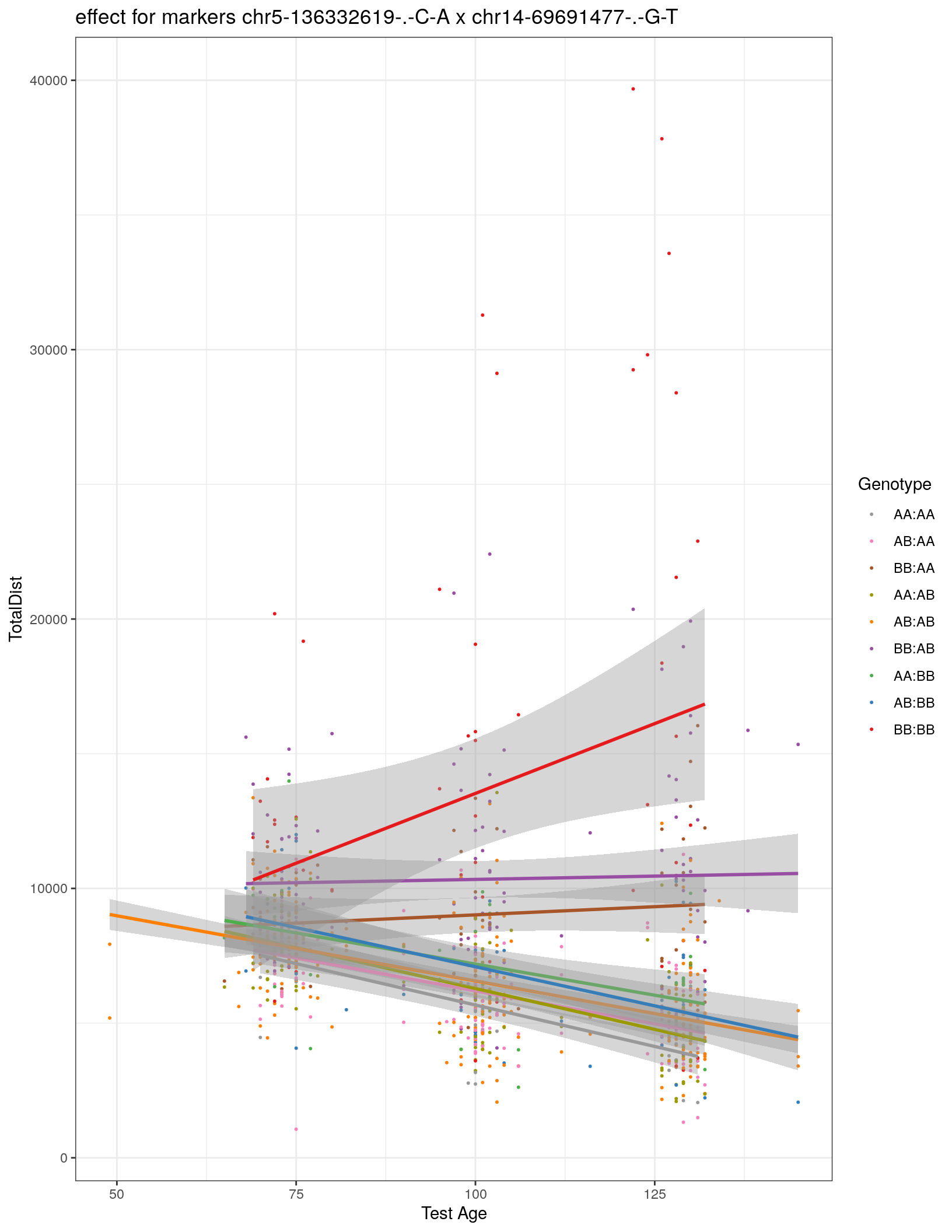
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
colnames(subplot)[colnames(subplot) == basem] = "Chr5"
colnames(subplot)[colnames(subplot) == mar] = "Chr14"
p2 <- ggplot(subplot, aes(Age, Dist, color=interactions, group=interactions)) +
geom_point(shape = 20, show.legend = FALSE) +
geom_line(aes(group = AnimalName, color = interactions), show.legend = FALSE, alpha = 0.2) +
geom_smooth(method='lm', formula= y~x, show.legend = FALSE, size = 1) +
#scale_color_discrete("Genotype") +
labs(color = "Genotype") +
ylab("Total distance traveled / Chr5") +
xlab("Test age / Chr14") +
#scale_color_brewer(palette = "Set1") +
scale_color_manual(breaks = c("AA:AA", "AB:AA", "BB:AA", "AA:AB",
"AB:AB", "BB:AB", "AA:BB", "AB:BB", "BB:BB"),
#values = RColorBrewer::brewer.pal(9, "Set1")[9:1]) +
values = c("#999999", "#F781BF", "#A65628","#9a9a00", "#FF7F00", "#984EA3", "#4DAF4A", "#377EB8", "#E41A1C")) +
stat_smooth(method="lm", show.legend = FALSE, formula = y~x) +
labs(title=paste0("Interaction effect between Chr5 and Chr14")) +
facet_grid(rows = vars(Chr5), cols = vars(Chr14), labeller = label_both) +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
theme(plot.title = element_text(hjust = 0.5))
print(p2)Warning: Removed 29 rows containing non-finite values (stat_smooth).Warning: Removed 29 rows containing non-finite values (stat_smooth).Warning: Removed 29 rows containing missing values (geom_point).Warning: Removed 29 row(s) containing missing values (geom_path).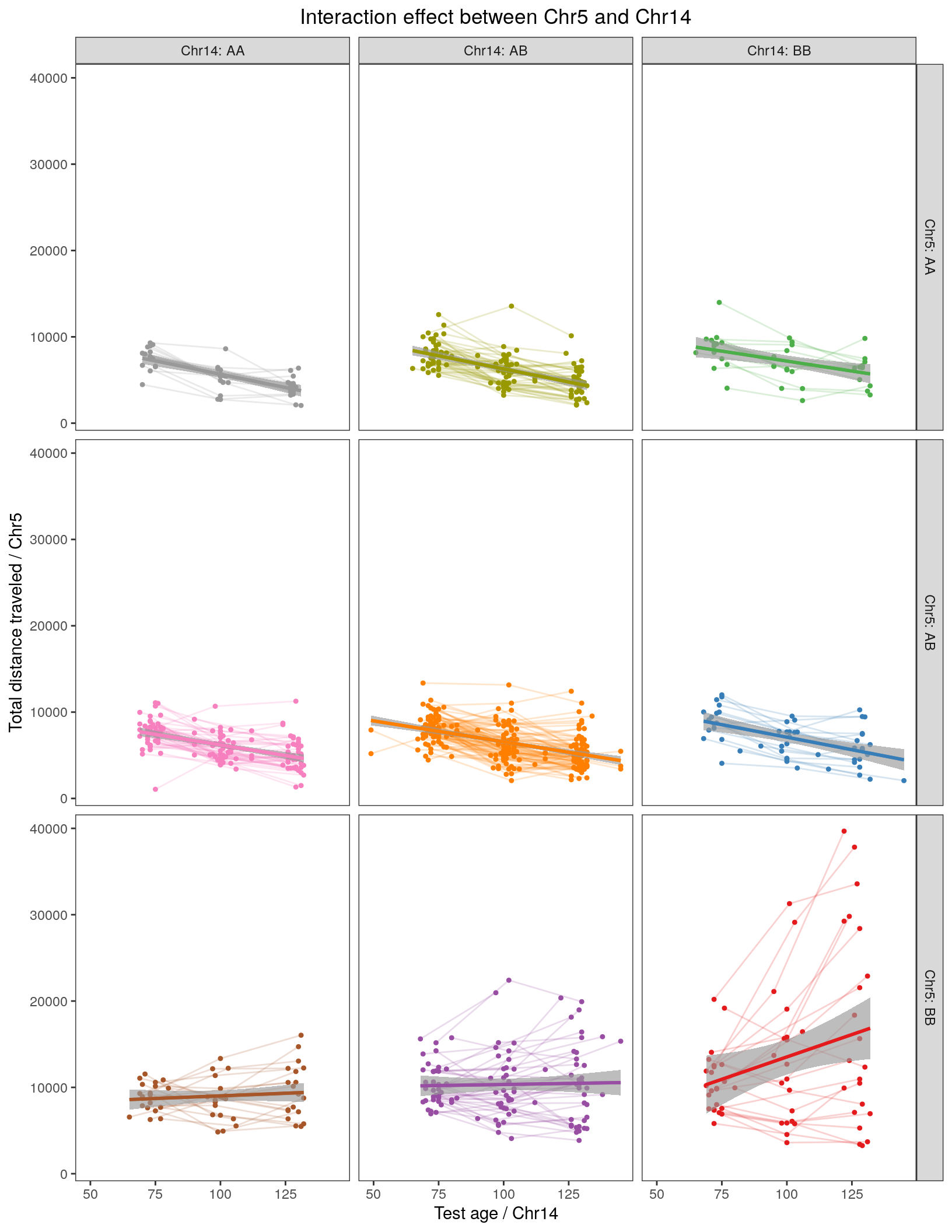
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
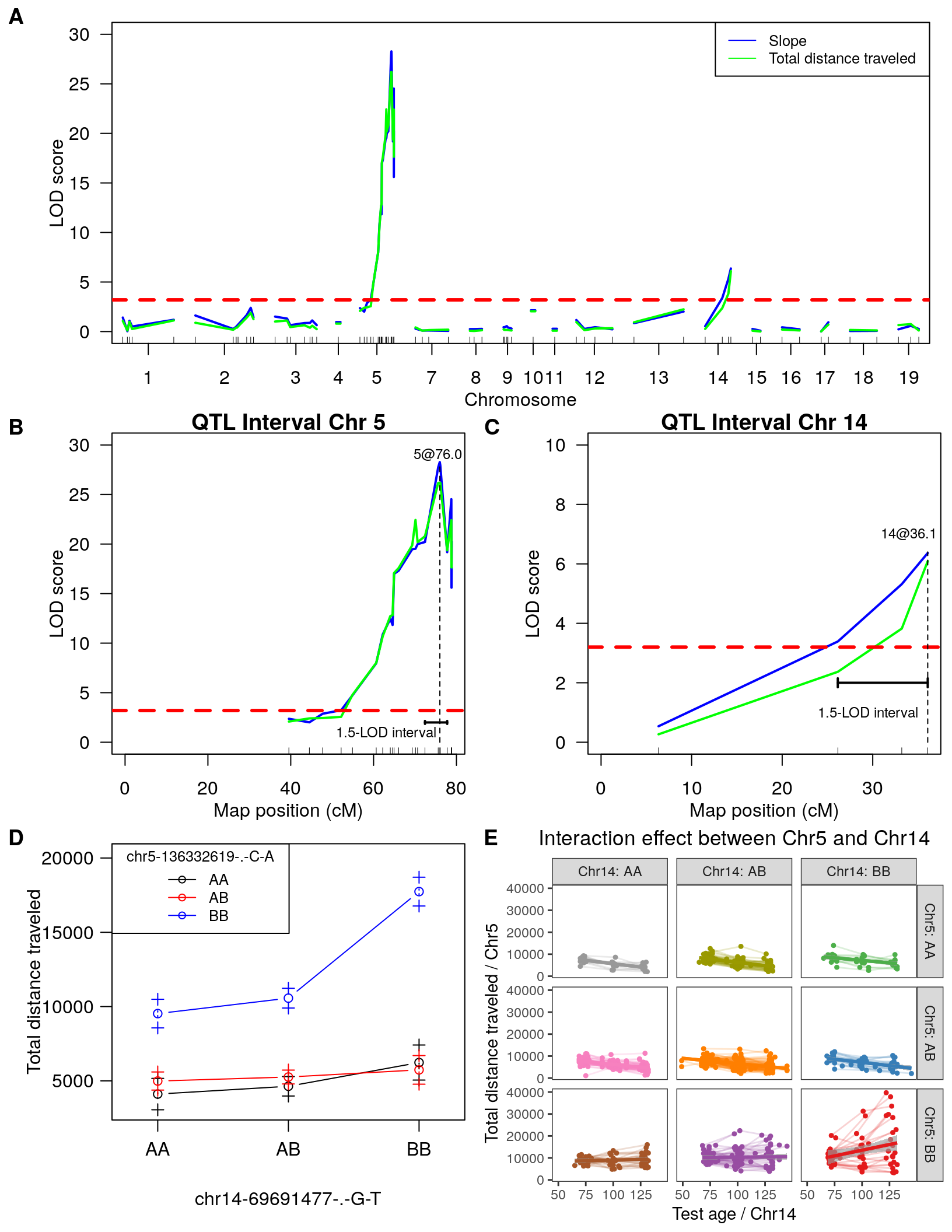
pp2 = plot_grid(figB, figC, labels = c("B", "C"), align = 'h', ncol=2)
pp3 = plot_grid(figD, p2, labels = c("D", "E"), align = 'h',ncol=2)Warning: Removed 29 rows containing non-finite values (stat_smooth).Warning: Removed 29 rows containing non-finite values (stat_smooth).Warning: Removed 29 rows containing missing values (geom_point).Warning: Removed 29 row(s) containing missing values (geom_path).Warning: Graphs cannot be horizontally aligned unless the axis parameter is set.
Placing graphs unaligned.pdf("output/figure_panel.pdf", width = 8.5, height = 11)
plot_grid(figA, labels = c('A', ''),
pp2,
pp3,
nrow = 3
)
dev.off()png
2 plot_grid(figA, labels = c('A', ''),
pp2,
pp3,
nrow = 3
)
| Version | Author | Date |
|---|---|---|
| 2470aab | xhyuo | 2022-12-24 |
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.2 LTS
Matrix products: default
BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 qtl2_0.24 lmerTest_3.1-3 lme4_1.1-26
[5] Matrix_1.3-2 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.4
[9] purrr_0.3.4 readr_1.4.0 tidyr_1.1.2 tibble_3.0.6
[13] tidyverse_1.3.0 qtlcharts_0.12-10 qtl_1.47-9 gridExtra_2.3
[17] ggplot2_3.3.3 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] nlme_3.1-152 fs_1.5.0 lubridate_1.7.9.2
[4] bit64_4.0.5 RColorBrewer_1.1-2 httr_1.4.2
[7] rprojroot_2.0.2 numDeriv_2016.8-1.1 tools_4.0.3
[10] backports_1.2.1 R6_2.5.0 mgcv_1.8-34
[13] DBI_1.1.1 colorspace_2.0-0 withr_2.4.1
[16] tidyselect_1.1.0 bit_4.0.4 compiler_4.0.3
[19] git2r_0.28.0 cli_2.3.0 rvest_0.3.6
[22] xml2_1.3.2 labeling_0.4.2 scales_1.1.1
[25] digest_0.6.27 minqa_1.2.4 rmarkdown_2.6
[28] pkgconfig_2.0.3 htmltools_0.5.1.1 highr_0.8
[31] dbplyr_2.1.0 fastmap_1.1.0 rlang_1.0.2
[34] readxl_1.3.1 rstudioapi_0.13 RSQLite_2.2.3
[37] gridGraphics_0.5-1 farver_2.0.3 generics_0.1.0
[40] jsonlite_1.7.2 magrittr_2.0.1 Rcpp_1.0.6
[43] munsell_0.5.0 lifecycle_1.0.0 stringi_1.5.3
[46] whisker_0.4 yaml_2.2.1 MASS_7.3-53.1
[49] grid_4.0.3 blob_1.2.1 parallel_4.0.3
[52] promises_1.2.0.1 crayon_1.4.1 lattice_0.20-41
[55] haven_2.3.1 splines_4.0.3 hms_1.0.0
[58] knitr_1.31 pillar_1.4.7 boot_1.3-27
[61] reprex_1.0.0 glue_1.4.2 evaluate_0.14
[64] data.table_1.13.6 modelr_0.1.8 vctrs_0.3.6
[67] nloptr_1.2.2.2 httpuv_1.5.5 cellranger_1.1.0
[70] gtable_0.3.0 assertthat_0.2.1 cachem_1.0.4
[73] xfun_0.21 broom_0.7.4 later_1.1.0.1
[76] memoise_2.0.0 statmod_1.4.35 ellipsis_0.3.1